Assays for detecting pathogens are used primarily to diagnose infections. Epidemiologists accumulate results from these tests in time series of case reports to conduct disease surveillance, a cornerstone of public health. During the COVID-19 pandemic, these data have been presented on dashboards of health agencies and media outlets all over the world. The shortcomings of these data have also become apparent: Trends can be misleading when demand for testing changes, when testing becomes more available, or when more (or less) accurate tests are rolled out. Time series of case counts are also a major simplification of the raw data used to generate them; modern diagnostics offer more than binary (positive or negative) results—they also estimate viral load, which can indicate the stage of infection. On page 299 of this issue, Hay et al. (1) develop an approach that uses aggregated viral load data to monitor epidemics more accurately than simple case series.
For most viruses, the contemporary standard assay for detection is quantitative (or real-time) polymerase chain reaction (qPCR). The number of cycles of the reaction at which an amplicon is at sufficient levels to produce a detectable signal is the cycle threshold (Ct) value. Because higher viral loads produce a signal at a lower number of reaction cycles, the Ct is inversely proportional to the amount of virus in the sample. Because acute viral infections follow a pattern whereby viral load peaks days to weeks after exposure and then declines, Ct values from qPCR can give an indication of the stage of an individual’s infection. A low Ct indicates high viral load and therefore the acute phase of illness; high Ct values (i.e., lower viral loads) occur during convalescence. But because viral loads are also low when an infection is just starting and are heterogeneous across individuals, a Ct value is typically not useful for informing an individual’s treatment.
However, more can be learned with Ct values at the population level. To understand the approach, consider an endemic infection where, on average, each case infects exactly one more person. Any snapshot in time would show a stable average viral load because some people are at the beginning of their illness and some are toward the end. It follows that, in a growing epidemic, more cases will be at the acute phase of illness and in a declining epidemic, more will be at a later phase, giving high and low average viral loads, respectively, at the population level (see the figure). This is the premise on which Hay et al. calculate the time-varying reproductive number (Rt) for COVID-19.
Viral load can be estimated from quantitative polymerase chain reaction (qPCR) testing for viral genomes. Aggregating viral load for a population can more reliably measure outbreak dynamics than case counts.
GRAPHIC: H. BISHOP/SCIENCE
” data-icon-position=”” data-hide-link-title=”0″>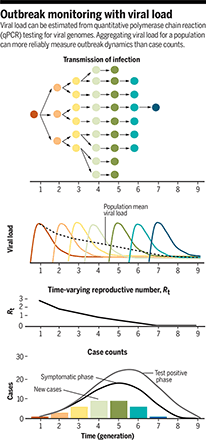

Viral load can be estimated from quantitative polymerase chain reaction (qPCR) testing for viral genomes. Aggregating viral load for a population can more reliably measure outbreak dynamics than case counts.
GRAPHIC: H. BISHOP/SCIENCE
Pathogen quantification by qPCR has been leveraged for various aspects of infectious disease epidemiology. Incorporating pathogen quantities has improved the ability to attribute infectious etiologies. This is not trivial for diseases that can be caused by more than one pathogen, especially when they often cause asymptomatic infections. This is most challenging in high-incidence settings where multiple pathogens are frequently detected in clinical samples. For example, because the more than 20 pathogens that cause diarrhea among children in low-resource settings are also frequently carried in the absence of diarrhea, detection of a pathogen in a diarrheal stool is not sufficient to assign etiology. But because the association with diarrhea increases with pathogen quantity for many enteric pathogens, statistical models that compare the quantity of pathogen detected by qPCR between diarrhea cases and controls can be used to estimate the population-level proportion of episodes that are attributable to each pathogen (2). Analogous applications have been used to attribute etiologies of severe pneumonia (3) and acute febrile illness to malaria (4).
Population-aggregated pathogen quantities have also played a role in controlling HIV. Virally suppressed individuals on antiretroviral therapy rarely transmit (5), so the community viral load has been used to quantify risk of HIV transmission and monitor test-and-treat control strategies. Community viral load, often calculated as the mean viral load of all infected individuals in a specific time and place, can correlate with HIV incidence and has predictable dynamics based on the characteristics of the HIV epidemic (6, 7).
Surveillance of pathogen quantity in the environment has also been used to track disease burden and interventions. Now used for COVID-19 (8), environmental surveillance has been important in efforts to eradicate polio (9). Poliovirus detected in sewage provides early warning for reintroduction of the virus in communities before cases of acute flaccid paralysis occur, and the quantity detected has been used to estimate the local prevalence of infections (10). Viral quantity in sewage was used to reconstruct a silent poliovirus outbreak in Israel as well as the impact of the oral polio vaccine, which is an attenuated poliovirus that is shed in stool and can be transmitted (11). In each of these examples, population-level aggregates of pathogen quantity have advanced understanding of disease burden and transmission.
The approach of Hay et al. to use pathogen quantification from cross-sectional surveys provides a rapid, efficient way to track disease dynamics. There is particular value in applying this method where testing capacity is limited. Random sampling could be resource-saving and more informative than indiscriminate testing of symptomatic individuals. There are, however, several limitations and some practical barriers. One is that there are infections for which the Ct is an unreliable indicator of the stage of infection (e.g., HIV) and/or symptom status (e.g., norovirus) (12). In addition, Ct values are only a proxy for pathogen quantities. Variability in laboratory protocols and specimen sampling procedures generates noise in pathogen load measurements. Furthermore, although widely used qPCR platforms are more standardized, commercially available kits tend to obscure the quantification process and only report the binary results. Replacing, or at least supplementing, symptom-based testing with structured random sampling will require a paradigm shift in how the use of qPCR is conceptualized at public health laboratories. Nonetheless, harnessing this information—already generated in massive quantities in diagnostic labs the world over—can be an important tool in monitoring the COVID-19 pandemic and future emerging infections.

