Polyclonal expansion of TCR Vbeta 21.3+ CD4+ and CD8+ T cells is a hallmark of Multisystem Inflammatory Syndrome in Children
Abstract
Multiple Inflammatory Syndrome in Children (MIS-C) is a delayed and severe complication of SARS-CoV-2 infection that strikes previously healthy children. As MIS-C combines clinical features of Kawasaki disease and Toxic Shock Syndrome (TSS), we aimed to compare the immunological profile of pediatric patients with these different conditions. We analyzed blood cytokine expression, and the T cell repertoire and phenotype in 36 MIS-C cases, which were compared to 16 KD, 58 TSS, and 42 COVID-19 cases. We observed an increase of serum inflammatory cytokines (IL-6, IL-10, IL-18, TNF-α, IFNγ, CD25s, MCP1, IL-1RA) in MIS-C, TSS and KD, contrasting with low expression of HLA-DR in monocytes. We detected a specific expansion of activated T cells expressing the Vβ21.3 T cell receptor β chain variable region in both CD4 and CD8 subsets in 75% of MIS-C patients and not in any patient with TSS, KD, or acute COVID-19; this correlated with the cytokine storm detected. The T cell repertoire returned to baseline within weeks after MIS-C resolution. Vβ21.3+ T cells from MIS-C patients expressed high levels of HLA-DR, CD38 and CX3CR1 but had weak responses to SARS-CoV-2 peptides in vitro. Consistently, the T cell expansion was not associated with specific classical HLA alleles. Thus, our data suggested that MIS-C is characterized by a polyclonal Vβ21.3 T cell expansion not directed against SARS-CoV-2 antigenic peptides, which is not seen in KD, TSS and acute COVID-19.
INTRODUCTION
At the end of April 2020, European clinicians warned the Public Health Agencies about an abnormal increase of Kawasaki-like diseases and myocarditis requiring critical care support in the context of the ongoing COVID-19 epidemic in children (1–3). American clinicians also reported a large outbreak of severe inflammation in children following COVID-19 infection, a condition that is now named Pediatric Inflammatory Multisystemic Syndrome (PIMS) or Multisystem Inflammatory Syndrome in children (MIS-C) (4–6). The clinical phenotype of this emerging disease is broad and encompasses features of Kawasaki disease (KD) and toxic shock syndrome (TSS). Many cases require intensive care support, making MIS-C one of the most severe manifestation of COVID-19 in children. Of note, MIS-C occurs 3 to 4 weeks after acute COVID-19 in children (3, 5–7).
To date, reports on MIS-C show slight differences in cytokine profiling and immunophenotype between MIS-C and KD or pediatric COVID-19 (8, 9). Analysis of T cells reveals a lower number of T cells in MIS-C with no or subtle signs of activation (10). Multi-dimensional immune profiling on small numbers of patients shows differences between acute COVID-19 or pre-pandemic KD (8, 11). A subset of activated CD8 T cells expressing the CX3C chemokine receptor (CX3CR1) is observed in MIS-C(12) and both CD8 and NK cells demonstrate an elevated expression of cytotoxicity genes (13). Anti-SARS-CoV2 antibodies are equally produced in pediatric COVID-19 and MIS-C. Autoantibodies are uniquely found during MIS-C or KD, which supports the contribution of the humoral response to both diseases (8, 11). Finally, a role for genetic factors is evocated in MIS-C pathogenesis as it occurs more frequently in Hispanic or African children (14–16). Despite these pioneer studies, the immunological mechanism underlying MIS-C remains unknown.
To address this question, we compared the immune profile in MIS-C patients to that of COVID-19 patients and that of patients with other clinically similar entities such as KD and TSS. For this, we explored the cytokine and cellular immune profile using different techniques. Using flow cytometry and transcriptomic analyses, we uncovered a specific Vβ21.3+ T cell expansion in 24/32 tested patients in MIS-C patients when assessed in the first month after onset. TCR sequencing revealed the polyclonal nature of the Vβ21.3+ expansion. No specific HLA bias was identified in patients, but we found a specific activation profile within Vβ21.3+ T cells. This activation was transient with a normalization of the repertoire within days to weeks after the inflammatory episode.
Together, our findings provide an immunological signature in MIS-C with potential implication in the diagnosis and treatment of this rare disease.
RESULTS
MIS-C presentation overlapped with TSS and KD
We took a cohort of 36 children with MIS-C and compared them with 16 KD cases diagnosed during and before the pandemic, 58 retrospective cases of TSS patients and 42 patients with acute COVID-19 (11 children, 31 adults). This comparison was motivated by previous descriptions of MIS-C in Europe and in the US, showing a clinical overlap between staphylococcal toxin-mediated TSS and KD in patients with MIS-C(1–3). Figure 1A outlines the study flowchart and the clinical and biological parameters we evaluated. Patients diagnosed for MIS-C, classical KD, TSS or acute COVID-19 were included. Patients were then subjected to deep immunological analyses combining cytokine profiling, TCR Vβ analysis and T cell stimulation assays (Fig. 1A). We confirmed the strong clinical overlap between MIS-C, TSS and KD. Indeed, many patients in the MIS-C group also fulfilled some of the 5 major criteria for TSS and KD respectively (Fig. 1B). Considering the clinical parameters, the most frequent features of MIS-C patients in our cohort were fever, cardiac dysfunction, gastrointestinal symptoms, coagulopathy and systemic inflammation (Table S1). Additional clinical data are presented in Table S2 for KD, TSS and acute COVID-19., and in Table S3 for all patients. Moreover, Table S4 gives a list of the patients analyzed in each of the following figure panels.
(A) Outline of the study including MIS-C, KD, TSS and acute COVID-19 patients and the immunological investigation workflow. (B) Heatmap showing the number of major criteria of TSS or KD for TSS, MIS-C and KD patients included in our study, considering the following case definition criteria: 5 clinical items for TSS (fever, rash, desquamation, hypotension, multisystem involvement) and 5 clinical items for KD in addition to fever (rash, cervical lymphadenopathy, bilateral conjunctivitis, oral mucosal changes, peripheral extremity changes).
” data-icon-position=”” data-hide-link-title=”0″>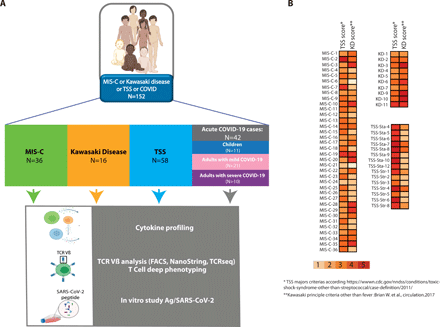
(A) Outline of the study including MIS-C, KD, TSS and acute COVID-19 patients and the immunological investigation workflow. (B) Heatmap showing the number of major criteria of TSS or KD for TSS, MIS-C and KD patients included in our study, considering the following case definition criteria: 5 clinical items for TSS (fever, rash, desquamation, hypotension, multisystem involvement) and 5 clinical items for KD in addition to fever (rash, cervical lymphadenopathy, bilateral conjunctivitis, oral mucosal changes, peripheral extremity changes).
High levels of proinflammatory cytokines in MIS-C contrasted with lymphopenia and low HLA-DR expression in monocytes.
SARS-CoV2 can cause fatal acute respiratory distress syndrome in patients at risk. This manifestation is caused by delayed and poorly controlled immune responses, with a deleterious role of inflammatory cytokines. Moreover, we and others have identified a subgroup of severe COVID-19 patients with impaired type-I interferon production (17–20). Thus, a regulated production of cytokines is paramount for control of SARS-CoV2 infection. This prompted us to investigate how cytokines could contribute to MIS-C pathogenesis. We compared the serum level of IFN-α, IFN-γ, TNF-α, IL-10, soluble CD25 (sCD25), MCP1, IL1Ra, IL-6 and IL-18 between healthy controls and MIS-C, KD, TSS and different forms of COVID-19 (mild pediatric, mild or severe adult-onset COVID-19, see Table S2 for a list of clinical features in the different patients’ groups).
The expression of interferon-stimulated genes (ISGs) in blood cells was significantly higher in MIS-C compared to controls, but rather low compared to COVID-19 patients (Fig. 2A-C). The level of serum IFNα2 followed the same trends, while serum IFNγ was variable among MIS-C patients, with very high levels in a few patients. The expression of the other cytokines measured (IL-6, IL-10, IL-18, TNFα, MCP1, IL1RA, CD25s) was very high in MIS-C patients compared to controls, and very similar to that of KD, TSS and severe COVID-19 patients (Fig. 2B-C). Of note the level of CD25s was significantly higher in TSS than in MIS-C patients, and significantly lower in severe COVID-19 patients than in MIS-C patients (Fig. 2B-C). A previous study found higher levels of serum IL-6 in KD patients than in MIS-C contrasting with our data (8).
(A) Left panel: Interferon score calculated as the normalized mean expression of six ISGs measured using the Nanostring technology, as previously described (44, 50). Middle panel: Serum IFN-α, in different groups of patients, as measured with the Simoa technology. Right panel: Serum IFN-γ level measured by Elisa. (B). Serum levels of the indicated cytokines as measured by automated ELISA. (C) Table showing the statistical results of the comparison of cytokine levels between MIS-C and other groups, as indicated. (D) T, B and NK lymphocyte counts measured by flow cytometry in MIS-C and KD. (E) HLA-DR expression in T cells and monocytes, as measured by flow cytometry in MIS-C. Gray shading indicates the derived central 95% HD reference interval (DE). See Table S4 for subject numbers per panel. Statistical test: Kruskal-Wallis test between healthy donors and all other groups with adjustment for multiple comparisons using Benjamini-Hochberg correction (AB) or between MIS-C and all other groups (C) with the same strategy. *P < 0.05, **P < 0.01, ***P < 0.001.
” data-icon-position=”” data-hide-link-title=”0″>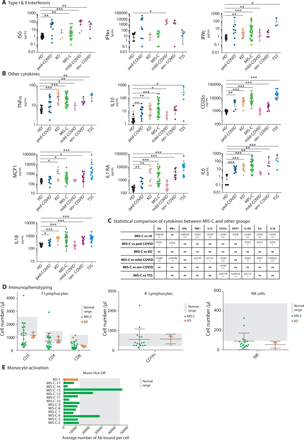
(A) Left panel: Interferon score calculated as the normalized mean expression of six ISGs measured using the Nanostring technology, as previously described (44, 50). Middle panel: Serum IFN-α, in different groups of patients, as measured with the Simoa technology. Right panel: Serum IFN-γ level measured by Elisa. (B). Serum levels of the indicated cytokines as measured by automated ELISA. (C) Table showing the statistical results of the comparison of cytokine levels between MIS-C and other groups, as indicated. (D) T, B and NK lymphocyte counts measured by flow cytometry in MIS-C and KD. (E) HLA-DR expression in T cells and monocytes, as measured by flow cytometry in MIS-C. Gray shading indicates the derived central 95% HD reference interval (DE). See Table S4 for subject numbers per panel. Statistical test: Kruskal-Wallis test between healthy donors and all other groups with adjustment for multiple comparisons using Benjamini-Hochberg correction (AB) or between MIS-C and all other groups (C) with the same strategy. *P < 0.05, **P < 0.01, ***P < 0.001.
To further explore the MIS-C immunological profile, we quantified the number of peripheral lymphocytes of different types, as well as the expression of HLA-DR in patient’s monocytes. T and NK cell counts were on average very low in MISC and KD patients while B cell counts were normal (Fig. 2D, S1). We found a decreased expression of HLA-DR in monocytes in both KD and MIS-C patients compared to controls (Fig. 2E, S1). Altogether our data show a strong similarity in cytokine profiles between MIS-C, KD and TSS and highlight the decreased lymphocyte counts and low HLA-DR expression in monocytes in MIS-C patients compared to controls.
Expansion of Vβ21.3+ peripheral T cells in a large fraction of MIS-C patients
TSST1-related TSS is associated with a skewing of the T cell repertoire toward Vβ2, as a result of TSST1-superantigen induced proliferation of Vβ2+ T cells (21). Every other S. aureus superantigenic toxin induces the expansion of specific TCR Vβ subsets, i.e., Vβ 5.2, 5.3, 7.2, 9, 16, 18, 22 for staphylococcal enterotoxin A (SEA) or Vβ 3, 12, 13.2, 14, 17, 20 for SEB (22). Given the similarities between TSS and MIS-C, we explored the possibility that MIS-C was also associated with specific T cell expansions. To explore the T cell repertoire in MIS-C, we first used flow cytometry to assess the distribution of Vβ subunits in T cells from MIS-C patients, in comparison with KD, TSS and COVID-19 patients (Fig. 3A, S2A). As expected, TSS patients displayed the hallmark expansion of the Vβ2+ subset. Interestingly, several Vβ-specific expansions were also visible in MIS-C patients, and in most cases Vβ21.3+ expansions (Fig. 3A), in both CD4 and CD8 T subsets (Fig. S3A-B). These expansions had similar amplitudes as the Vβ2+ expansions in TSS (Fig. 3A). A principal component analysis of the Vβ distribution in CD4 and CD8 T cells showed that the main parameters separating the different patients were the frequency of Vβ2+ and the frequency of Vβ21.3+ cells (Fig. S3C-D). Overall, the expansion of Vβ21.3+ T cell subsets was seen in 15/26 (58%) of MIS-C patients and in none of the other conditions analyzed by flow cytometry ie KD, TSS and COVID-19 (Fig. 3A). Next, we wanted to use a different technique to test the specificity of this expansion, and we therefore performed transcriptomic analyses of Vβ expression in PBMC using the Nanostring technology. This technique also requires much less material than flow cytometry, which allowed us to run lymphopenic samples from severe COVID-19 cases. This transcriptomic analysis firmly established that the Vβ21.3+ T cell expansion is a hallmark of MIS-C as it was seen in 18/23 MIS-C patients tested (Fig. S3D). Thus, taking together flow cytometry and Nanostring analyses, we found that 24/32 (75%) of MIS-C patients and none in the other clinical groups, displayed TRBV11-2/Vβ21.3+ expansions.
(A) Frequency of total CD3+ T cells expressing the indicated V-beta (Vβ) chains, as measured by flow cytometry using specific antibodies against the corresponding Vβ within PBMCs of patients of the indicated group. TSS, mild COVID-19, pediatric COVID-19 (ped-COVID), KD and MIS-C patients are colored in blue, pink, dark blue, orange and green respectively. Bubbles represent the normalized individual Vβ frequency reported to the mean frequency for each Vβ in the general adult population. (B) Normalized frequency of Vβ21.3+ T cells in different clinical conditions, as indicated. (C-D) Serum IL-18 (C) and IL-1RA (D) levels in MIS-C patients with or without Vβ21.3+ T cell expansions (exp). (E-G) Chord diagrams of the TRBV (bottom, grey) and TRBJ (top, blue) combinations assessed by TCR sequencing of TCRαβ chains in whole blood of MIS-C patients. The relative frequency of all TRBVBJ combinations have been calculated per sample on the full TRB repertoire data. Combinations using TRBV11-2 are highlighted in red. Each red line indicates pairing with a given TRBJ, the thickness indicates the frequency of this pairing. The percentage values under each chart indicate the percentage of clonotypes composed with the TRBV11-2 gene. In (E-G) the CDR3 length distribution of clonotypes using TRBV11-2 is shown as a histogram graph. Each clonotype is represented as a grey line. The thickness of the line represents the frequency of the clonotype within each repertoire. Since most of the clonotypes are not abundant, all the grey lines are stacked together and appear as a unique grey bar, which reflect the lack of expansion. Expanded clonotypes identified as detailed in the method section are shown in red. In (F-G) the same four patients are shown during the MIS-C episode (F) and after resolution (G). (H) Frequency of Vβ21.3+ T cells at different time points during and after the MIS-C episode in different patients, as assessed by flow cytometry. (I) Annexin-V staining of T cells in the indicated patients’ groups. Results show the ratio of the Annexin-V fluorescence in Vβ21.3+ vs Vβ21.3- T cells. See Table S4 for subject numbers per panel. (B) Statistical test: Kruskal-Wallis test between MIS-C and all other groups with adjustment for multiple comparisons using Benjamini-Hochberg correction, (CDI) unpaired Wilcoxon test comparing two groups. *P < 0.05, **P < 0.01, ***P < 0.001. ****P<0.0001
” data-icon-position=”” data-hide-link-title=”0″>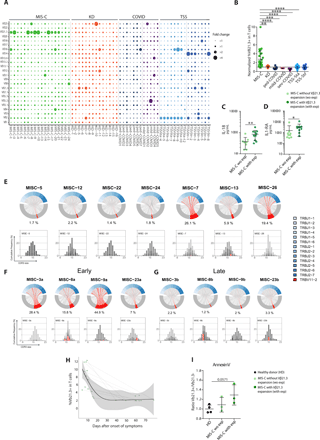
(A) Frequency of total CD3+ T cells expressing the indicated V-beta (Vβ) chains, as measured by flow cytometry using specific antibodies against the corresponding Vβ within PBMCs of patients of the indicated group. TSS, mild COVID-19, pediatric COVID-19 (ped-COVID), KD and MIS-C patients are colored in blue, pink, dark blue, orange and green respectively. Bubbles represent the normalized individual Vβ frequency reported to the mean frequency for each Vβ in the general adult population. (B) Normalized frequency of Vβ21.3+ T cells in different clinical conditions, as indicated. (C-D) Serum IL-18 (C) and IL-1RA (D) levels in MIS-C patients with or without Vβ21.3+ T cell expansions (exp). (E-G) Chord diagrams of the TRBV (bottom, grey) and TRBJ (top, blue) combinations assessed by TCR sequencing of TCRαβ chains in whole blood of MIS-C patients. The relative frequency of all TRBVBJ combinations have been calculated per sample on the full TRB repertoire data. Combinations using TRBV11-2 are highlighted in red. Each red line indicates pairing with a given TRBJ, the thickness indicates the frequency of this pairing. The percentage values under each chart indicate the percentage of clonotypes composed with the TRBV11-2 gene. In (E-G) the CDR3 length distribution of clonotypes using TRBV11-2 is shown as a histogram graph. Each clonotype is represented as a grey line. The thickness of the line represents the frequency of the clonotype within each repertoire. Since most of the clonotypes are not abundant, all the grey lines are stacked together and appear as a unique grey bar, which reflect the lack of expansion. Expanded clonotypes identified as detailed in the method section are shown in red. In (F-G) the same four patients are shown during the MIS-C episode (F) and after resolution (G). (H) Frequency of Vβ21.3+ T cells at different time points during and after the MIS-C episode in different patients, as assessed by flow cytometry. (I) Annexin-V staining of T cells in the indicated patients’ groups. Results show the ratio of the Annexin-V fluorescence in Vβ21.3+ vs Vβ21.3- T cells. See Table S4 for subject numbers per panel. (B) Statistical test: Kruskal-Wallis test between MIS-C and all other groups with adjustment for multiple comparisons using Benjamini-Hochberg correction, (CDI) unpaired Wilcoxon test comparing two groups. *P < 0.05, **P < 0.01, ***P < 0.001. ****P<0.0001
We then compared the level of serum cytokines between MIS-C patients with and without Vβ21.3+ T cell expansions, at the time of the acute episode. The levels of IL-18 and IL-1RA (Fig. 3C-D) were associated with the polyclonal Vβ21.3 expansions, but not those of the other cytokines tested (Fig. S4A-B), suggesting that Vβ21.3+ T cells were associated with the cytokine storm.
TCR sequencing highlighted the polyclonal nature of TCR Vβ21.3 expansions
To investigate the clonality of Vβ21.3+ expanded cells, we analyzed the TCR repertoire of 11 MIS-C patients for whom whole blood RNA was available by TCR-sequencing. We analyzed the composition of the TCR beta rearrangements involving the TRBV11-2 gene (which corresponds to Vβ21.3). First, by representing the TRBV11-2/TRBJ combination usage as chord diagrams (Fig. 3E, 3F), we confirmed the expansion of T cells using TRBV11-2 in 7 out of the 11 patients. These TRBV11-2 rearrangements were associated with multiple TRBJ genes, suggesting the polyclonal nature of the expansions. To further evaluate the polyclonality, we analyzed the hypervariable sequence CDR3 length distribution of TRBV11-2 clonotypes (barplots Fig. 3E-G.). The CDR3 size distributions showed a bell-shaped Gaussian distribution as expected in polyclonal repertoires (23–25). In order to evaluate the degree of polyclonality, we identified the expanded clonotypes by setting a threshold based on the binomial distribution of the clonotype frequencies per sample (see methods section and Fig. S5A). No major monoclonal expansions (red lines in the CDR3 spectratypes) explaining the global TRBV11-2 expansion were detected. Instead, most of the clonotypes were found at low frequencies (grey lines), typical of a polyclonal diverse repertoire. The percentages of expanded clonotypes were not significantly different between patients with or without TRBV11-2. We calculated the cumulative frequencies of such expanded clonotypes within the full repertoire and found that they were always far below the frequency of the full TRBV11-2 expansion in patients with expansions, representing in average 0.51% of the total repertoire. Finally, these limited expansions represented in average 4.47% of the TRBV11-2 repertoire in patients with TRBV11-2 expansions and 6.31% in patients without TRBV11-2 expansions (Table S6 and Fig. S5B). To confirm the polyclonality of the TRBV11-2 expansion, we computed the Berger-Parker index (BPI) on TRBV11-2 clonotype for MIS-C patients harboring or not TRBV11-2 expansions (Fig. S5C). This index measures the proportional abundance of the most frequent clonotypes within TRBV11-2 clonotypes. There were no significant differences when we compared the BPI on TRBV11-2 clonotypes between patients with or without TRBV11-2 expansions, further confirming that TRBV11-2 expansions in the 7 patients were not explained by monoclonal expansions.
Next, to address whether the Vβ21.3+ T cell expansion persisted overtime, we repeated the TCR sequencing and the flow cytometry Vβ analyses in a group of patients for which blood samples were available during and after the acute inflammatory episode. As shown in Figs. 3F-H, the Vb21.3/TRBV11-2 distributions for all the patients returned to normal within days to weeks after MIS-C. Interestingly, when we compared the CDR3 length distributions by calculating the perturbation score using the ISEApeaks tool between repertoires obtained during and after the acute response, we found no differences between the two groups, further supporting the polyclonal expansion profile of TRBV11-2 during the acute response (Fig. S5D). Finally, this transient expansion suggested a pro-apoptotic phenotype of Vβ21.3+ T cell. To test this hypothesis, we stained PBMCs from MIS-C patients with Annexin-V that marks early apoptotic cells. A higher fraction of Vβ21.3+ compared with Vβ21.3- T cells were stained with Annexin-V in MIS-C patients with Vβ21.3+ expansions (Fig. 3I, S2B), which substantiated our hypothesis.
Vβ21.3+ T cells had an activated phenotype but did not react against SARS-CoV2 peptides
As Vβ21.3+ T cells expand in MIS-C patients, we investigated their activation status and the mechanisms underlying their proliferation. We found that the activation markers HLA-DR and CD38 were expressed at high levels in both CD4 and CD8 T cells from MIS-C patients with Vβ21.3+ expansions compared to those without expansions and to healthy controls (Fig. 4A, 4B). This was due to a specific up regulation of CD38 and HLA-DR in Vβ21.3+ CD4 and CD8 T cells in MIS-C patients with Vβ21.3 expansions compared to those without Vβ21.3 expansions (Fig. 4C, 4D). A recent paper reports a specific activation of CX3CR1+ CD4 and CD8 T cells in MIS-C patients, as assessed by HLA-DR/CD38 levels (12). This prompted us to measure CX3CR1 levels in Vβ21.3+ T cells. As shown in Fig. 4E and Figure S6A, Vβ21.3+ T cells overexpressed CX3CR1 in both CD4 and CD8 T cells in MIS-C patients with Vβ21.3+ expansions compared to those without expansions, even though the percentage of CX3CR1 positive cells was not higher in MIS-C than in control patients (Fig. S7A). Moreover, in MIS-C patients, a large frequency of non-naïve CX3CR1+ CD4 and CD8 T cells had an activated phenotype as previously reported (12) (Fig. S7B).
(A-D) Flow cytometry analysis of CD38 and HLA-DR expression in CD4 or CD8 T cells from the indicated patients’ groups (exp: Vβ21.3+ T cell expansion, we = without). (A) shows a representative staining, and (B) shows the mean +/−SD frequency of CD38+HLA-DR+ CD4 (top) and CD8 (bottom) T cells. (C-D) A Vβ21.3+ antibody was also included in the flow cytometry panel used in (A-B) allowing a specific comparison of the Vβ21.3- and Vβ21.3+ T cells in MIS-C patients. (C) shows a representative dot plot of CD38 and HLA-DR expression in the indicated subsets; (D) mean +/−SD frequency of CD38+HLA-DR+ in the indicated CD4 (top) and CD8 (bottom) T cell subsets. (E) Frequency of CX3CR1+ cells in gated Vβ21.3- and Vβ21.3+ CD4+ (left) and CD8+ (right) T cells in MIS-C without and MIS-C with expansion. (F) PBMCs from control, COVID-19 (adults, 6 months post infection) or MIS-C patients (with or without Vβ21.3+ T cell expansions) were stimulated for 6h with a commercial cocktail of synthetic peptides from S, N, and M SARS-CoV2 proteins in the presence of Golgi secretion inhibitors. Intracellular IFNγ expression was then measured in T cells by flow cytometry. The fold increase was calculated as the ratio between the stimulated and the unstimulated conditions. (G) shows the frequency of Vβ21.3+ and Vβ21.3- T cells expressing IFN-γ after stimulation with S, N, M SARS-CoV2 peptides in the different patient groups as indicated (one dot: one patient). See Table S4 for subject numbers per panel (B) Kruskal-Wallis test between three groups with adjustment for multiple comparisons using Benjamini-Hochberg correction, (DEFG) Wilcoxon test comparing two groups. *P < 0.05.
” data-icon-position=”” data-hide-link-title=”0″>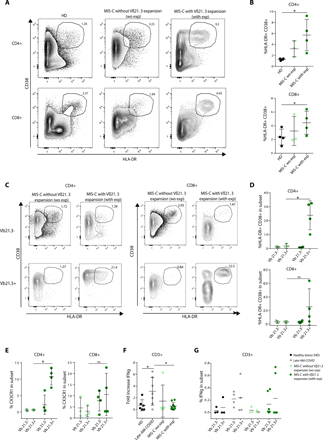
(A-D) Flow cytometry analysis of CD38 and HLA-DR expression in CD4 or CD8 T cells from the indicated patients’ groups (exp: Vβ21.3+ T cell expansion, we = without). (A) shows a representative staining, and (B) shows the mean +/−SD frequency of CD38+HLA-DR+ CD4 (top) and CD8 (bottom) T cells. (C-D) A Vβ21.3+ antibody was also included in the flow cytometry panel used in (A-B) allowing a specific comparison of the Vβ21.3- and Vβ21.3+ T cells in MIS-C patients. (C) shows a representative dot plot of CD38 and HLA-DR expression in the indicated subsets; (D) mean +/−SD frequency of CD38+HLA-DR+ in the indicated CD4 (top) and CD8 (bottom) T cell subsets. (E) Frequency of CX3CR1+ cells in gated Vβ21.3- and Vβ21.3+ CD4+ (left) and CD8+ (right) T cells in MIS-C without and MIS-C with expansion. (F) PBMCs from control, COVID-19 (adults, 6 months post infection) or MIS-C patients (with or without Vβ21.3+ T cell expansions) were stimulated for 6h with a commercial cocktail of synthetic peptides from S, N, and M SARS-CoV2 proteins in the presence of Golgi secretion inhibitors. Intracellular IFNγ expression was then measured in T cells by flow cytometry. The fold increase was calculated as the ratio between the stimulated and the unstimulated conditions. (G) shows the frequency of Vβ21.3+ and Vβ21.3- T cells expressing IFN-γ after stimulation with S, N, M SARS-CoV2 peptides in the different patient groups as indicated (one dot: one patient). See Table S4 for subject numbers per panel (B) Kruskal-Wallis test between three groups with adjustment for multiple comparisons using Benjamini-Hochberg correction, (DEFG) Wilcoxon test comparing two groups. *P < 0.05.
Given that MIS-C came about weeks after COVID-19, we wondered if Vβ21.3+ T cells were raised against SARS-CoV-2 antigens. To test this possibility, we stimulated PBMCs from MIS-C or convalescent COVID-19 patients with a commercial cocktail of SARS-CoV2 peptides spanning S, N and M viral proteins. T cells from MIS-C patients responded poorly to stimulation with viral peptides, regardless of Vβ21.3 expansion, compared to T cells from convalescent COVID-19 patients that responded well (Fig. 4F, 4G, S6B-C). This was not due to a lack of adaptive anti-SARS-CoV-2 response, because all MIS-C patients tested had high SARS-CoV-2-specific antibody levels (Fig. S7C-E). Finally, we could not identify any specific allele nor mutations of classical HLA class I or class II genes associated with TRBV11-2 expansions by genomic sequencing of the HLA loci of 13 MIS-C patients (Table S4). Together with the lack of Vβ21.3+ expansion in COVID-19 patients, these data showed that Vβ21.3+ T cells were not specific for HLA-restricted SARS-CoV-2 peptides.
Together, these data revealed that the Vβ21.3+ CD4 and CD8 T cell expansion were highly activated and expressed CX3CR1, but had poor responsiveness to SARS-CoV-2 antigens.
DISCUSSION
Here, we confirmed the strong overlap in clinical phenotype between KD, MIS-C, and TSS; MIS-C and TSS had similar defining features, specifically cardiac dysfunction, hypotension, maculo-papular skin rash and conjunctivitis. We recently identified the critical importance of early steroid therapy in the management of MIS-C, similarly to what has been previously shown in TSS (26, 27). MIS-C and TSS are obviously linked to infections, while many KD features suggest an infectious cause for KD as well (28). The epidemic of a novel coronavirus in 2005 (New Haven) was associated to KD and linked the viral infection to vascular inflammation (29).
We found important similarities in terms of cytokine expression between MIS-C, TSS and KD such as high TNF-α, IL6, IL18 and IL1Ra levels. A previous study noted that a subgroup of severe MIS-C patients had higher levels of IFN-γ, IL-18, GM-CSF, RANTES, IP-10, IL-1α, and SDF-1 than mild MISC or KD patients (30). We also observed a subset of MIS-C patients with high serum IFN-γ, IL-18 and CD25s. These observations confirm previous reports showing a clinical and biological overlap between MIS-C and macrophage activation syndrome (3), and suggest the importance of IFN-γ in the disease.
Here, we reported the expansion of a TCR Vβ21.3+ T cell subset with an activated phenotype in as many as 75% of MIS-C patients. Vβ21.3+ T cell expansions were also reported in smaller numbers of MIS-C patients in two recent studies (13, 37). In both Porritt et al. and our study Vβ21.3+ T cell expansions appeared polyclonal as judged by the large number of TRBJ gene segments associated with TRBV11.2 and by the even distribution of the CDR3 domain. Our study further showed that Vβ21.3+ CD4 and CD8 T cell expansions are a discriminating feature of MIS-C compared to KD, TSS and COVID-19 patients.
We observed a correlation between Vβ21.3+ T cell expansions and the level of serum cytokines IL-18 and IL-1RA from matching samples, confirming a previous study (37) and indicating that Vβ21.3+ T cell expansions were associated with the cytokine storm. Our data also showed that Vβ21.3+ T cells have an activated phenotype, with high HLA-DR and CD38 expression and that activated Vβ21.3+ T cells expressed high levels of CX3CR1, a marker of patrolling monocytes and of cytotoxic lymphocytes. CX3CR1 binds to CX3CL1, a membrane-bound chemokine induced on vascular endothelial cells upon inflammation (12). The CX3CL1-CX3CR1 axis is thought to have an important role in vascular inflammation in different inflammatory diseases (38), and could contribute to MIS-C pathogenesis. This interaction could promote the cytotoxic action of different lymphocyte populations, which fits with the reported elevated expression of cytotoxicity genes in NK and CD8+ T cells in MIS-C patients (13).
We demonstrated that both TSS and MIS-C were marked by the polyclonal proliferation of a specific Vβ subset i.e., Vβ2+ cells for TSS related to TSST1, and Vβ21.3+ cells for MIS-C. The amplitude of the expansion was also similar in both syndromes. Considering the other clinical phenotype similarities between MIS-C and TSS shown in this study, cytokine production and treatment, this raises the hypothesis that Vβ21.3+ cell expansions are caused by a superantigen structure in MIS-C. The term superantigen has been coined by Kappler and Marrack as an operational definition of various T-cell activating substances with specificity for T cell antigen receptors Vβ subunits regardless of the rearrangement and antigen-specificity (39). Superantigens bind external regions of T cell receptor and MHC molecules (40) and can induce massive expansions of T cells expressing one specific TCR Vβ chain while classical antigens induce the expansion of T cells bearing different Vβ. Previous papers have suggested that the SARS-CoV2 Spike protein could behave as a superantigen structure (41). Using in silico modelling, Porritt et al. identified a putative interaction between Vβ21.3 and a superantigen-like motif in Spike. However, Vβ21.3+ T cell expansions occur in a delayed manner relative to SARS-CoV-2 infection, and the virus is often undetectable in MIS-C patients at the time of the acute inflammation. The kinetics of MIS-C relative to COVID-19 is compatible with a causal role of anti-SARS-CoV-2 antibodies. One can hypothesize that immune complexes composed of SARS-CoV-2 bound to antibodies may act as superantigen structures. However, a previous study failed to detect such immune complexes in MIS-C patients (30). In addition, Vβ restricted T cells adhere to endothelial cells following superantigen activation (42) and thus the CX3CR1+ Vβ21.3 expanded T cells may play a role in vascular injury in MIS-C. Alternative mechanisms may be put forward, such as secondary autoimmune reactions. Several studies have indeed reported the appearance of autoantibodies in MIS-C patients, some of which directed against endothelial antigens (8, 11), while others have reported immune events consistent with autoimmunity such as the expansion of proliferating plasmablasts (13) or the persistence of functional SARS-CoV-2-specific monocyte-activating antibodies (43). How B cell mediated autoimmunity would be linked to Vβ specific T cell expansions is however unclear. One could speculate that immune complexes composed of autoantibodies and endogenous antigens could behave as superantigens.
Finally, given the rarity of MIS-C, there could be a genetic susceptibility to this post-infectious disease promoting hyperinflammatory reaction of adaptive immunity in response to SARS-CoV2 (16). We limited our analysis to classical HLA alleles, but did not find any significant association, even though a previous study reported an HLA-I bias in a smallest group of MIS-C patients (37). Of note our MIS-C samples were obtained in most of cases after anti-inflammatory treatments (see supplementary Table 1), and it is likely that those treatments affect the level of serum cytokines, which could have impacted the comparisons we made between clinical conditions, and the associations between cytokines and T cell expansions. Taken together, MIS-C shared clinical and immunological anomalies with KD and TSS, but was specifically characterized by a polyclonal Vβ21.3 expansion in CD4 and CD8 T cells associated to activation and CX3CR1 expression.
MATERIALS AND METHODS
More information for all of these protocols can be found in the supplementary materials.
Study design and human subjects
The immunological profile of 36 MIS-C cases, 16 KD and 58 TSS cases and 42 non-MISC COVID-19 were included (Fig. 1A). Samples were collected within the first week of symptoms and analyzed for cytokine immunoprofiling, standard immunophenotyping, Vβ expression, TCR sequencing, SARS-CoV2-dependent T cell response. Because of low volume sampling of pediatric patients, we did not have the same availability for research blood draws. The samples used for each experiment are detailed in Table S4. The main clinical features are summarized in Table S1, S2, S3. Written informed consent was obtained for all data collection and blood sampling as detailed in supplementary material.
Immunological analyses
Cytokines and IFN score assessment
Plasma concentrations of IL-6, TNF-a, IFN-g, IL-10, IL-18, MCP-1, IL-1ra and CD25s were measured by Simpleplex technology using ELLA instrument (ProteinSimple). Plasma IFN-α concentrations were determined by single-molecule array (Simoa) on a HD-1 Analyzer (Quanterix) using a commercial kit for IFN-α2 quantification (Quanterix). RNA was extracted from whole blood and IFN score was obtained using nCounter analysis technology (NanoString Technologies) by calculating the median of the normalized count of 6 ISGs as previously described (44)
T-cell Vβ repertoire analysis and immunophenotyping
PBMCs were stained with surface markers, CD3, CD4, CD8, CD14, CD16, CD19, CCR7, CD38, Vb21.3, HLA-DR, CX3CR1, CD45RA (further details on these stains are included in the supplemental materials). Cell apoptosis was assessed by annexin V staining. All samples were acquired on a BD LSR Fortessa (BD Biosciences) flow cytometer and analyzed using FlowJo version 10 software. Monocyte HLA-DR expression was determined on EDTA-anticoagulated peripheral whole blood as previously described (45). The phenotypic analysis of T-cell Vβ repertoire was performed on whole blood sample using the IOTest Beta Mark kit (Beckman-Coulter). Whole blood cells were stained for CD3, CD4, CD8 and each combination of 3 FITC-, PE- and FITC/PE-conjugated anti-Vβ mAbs (Beckman-Coulter) in 8 sample tubes. Expansions were defined respectively for values above the mean+2SD or below the minimum reference values of the corresponding family. Additional samples were analyzed for TRBV from total RNA with Nanostring technology (Supplemental Material).
TCR-sequencing and analysis
T cell receptor (TCR) alpha/beta libraries were prepared from 300ng of RNA from each sample with SMARTer Human TCR a/b Profiling Kit (Takarabio) (32). The sequencing was carried out on a MiSeq Illumina sequencer using the MiSeq v3 PE300 protocol at the Biomics Platform (Institut Pasteur, Paris, France). Single end sequences were aligned and annotated using MiXCR 3.0.13 (46), providing a list of clonotypes. Analyses were performed in R 4.0.3 on the TRB clonotype lists obtained with MiXCR. For each clonotype, read count was recorded. Frequencies for TRBV, TRBJ and clonotypes were calculated based on the total read counts per sample. Chord diagrams were made using the circlize package (47) on TRBVBJ frequencies, CDR3 length spectratypes were made using ggplot2 (48) using clonotype frequencies. To identify TRBV11-2 expanded clonotypes, first (Q1) and third (Q3) quartiles and the interquartile range (IQR) were computed for all patients without TRBV11-2 expansion. Expanded clonotypes are defined as those with counts superiors to Q3+(1.5*IQR).
Stimulation with SARS-CoV-2 overlapping peptide pools and flow cytometry
PBMCs were stimulated with SARS-CoV-2 PepTivator pooled S, N, M peptides (Miltenyi Biotec) at a final concentration of 2 μg ml−1 for 1 h in the presence of 2 μg ml−1 monoclonal antibodies CD28 and CD49d, and then for an additional 5 h with GolgiPlug and GolgiStop (BD Biosciences). Similar surface markers were stained. Cells were then washed, fixed with Cytofix/Cytoperm (BD Biosciences) and stained with V450- conjugated anti-IFNγ. All samples were acquired on a BD LSRFortessa (BD Biosciences) flow cytometer and analyzed using FlowJo version 10 software.
Serology
Serum samples were tested with three commercial assays: the Wantai Ab assay detecting total antibodies against the receptor binding domain (RBD) of the S protein, the bioMérieux Vidas assay detecting IgG to the RBD and the Abbott Architect assay detecting IgG to the N protein.
Statistical analyses
All tests were performed two-sided with a nominal significance threshold of p<0.05. We used nonparametric tests appropriated to the low number of observations in each of our experimental conditions, i.e., the Wilcoxon or Kruskal-Wallis test depending on whether we have 2 or more conditions to compare, respectively. Multiple comparisons performed with the Dunn's all-pairs comparison for Kruskal-type ranked data were corrected by the fdr method of Benjamini-Hochberg (49). PCA analysis was made in R with stats package and visualized with ggplot2 (48) for Vbeta frequencies obtained by flow cytometry. All statistical analyses were performed using GraphPad with the help of a professional biostatistician.
Supplementary Materials
immunology.sciencemag.org/cgi/content/full/6/59/eabh1516/DC1
Fig. S1: Assessment of T, B, NK cells and HLA-DR monocytes
Fig. S2: Assessment of T-cell receptor repertoire and T cell apoptosis by flow cytometry
Fig. S3: Vβ TCR repertoire analysis
Fig.S4: Cytokine assessment in MIS-C
Fig. S5: TRBV11-2 polyclonality assessment
Fig. S6: Analysis T cell activation within Vβ21.3 by flow cytometry
Fig. S7: T cell phenotyping and SARS-CoV2 serology
Table S1: MIS-C patients clinical characteristics
Table S2: non MIS-C patients clinical characteristics
Table S3: Individual data of all included patients
Table S4: List of patients analysed per figure panel
Table S5: HLA sequencing in 13 MIS-C patients
Table S6: TRBV11-2 clonotype expansions
Table S7: Raw Data File (excel spreadsheet)
- Copyright © 2021, American Association for the Advancement of Science


