Integrated longitudinal immunophenotypic, transcriptional and repertoire analyses delineate immune responses in COVID-19 patients
RESULTS
Broad immune remodelling in COVID-19 patients
To assess the dynamics of immune responses elicited by SARS-CoV-2 infection, we collected PBMCs from COVID-19 patients at the time of acute infection (hereafter indicated as “infection”), namely within 21 days from the diagnosis, and weeks after the resolution of the infection (hereupon “post-infection”), demonstrated by negative nasopharyngeal swab, following a previous positivity. We investigated innate and adaptive immune responses in 17 patients, 6 with mild disease (no interstitial pneumonia, no oxygen requirement) and 11 with severe disease (pneumonia with respiratory failure), and compared them to 4 healthy individuals (HD). Demographic and clinical characteristics of patients are shown in Table S1. The median age of patients was 55 years (IQR 39-70), 7/17 (41.2%) patients were females and 11/17 (64.7%) had one or more co-morbidities. In patients with pneumonia requiring oxygen support, the median PaO2:FiO2 ratio at the time of hospital admission was 200 mmHg. Lymphopenia (<1×109/L lymphocytes) was registered at time of blood collection in 6/15 patients (40%; 2/17 n/a). All patients with mild disease were under 50 years of age and 2/6 (33.33%) of them had co-morbidities. 9/11 (81.81%) of patients with severe disease were over 50 years of age (5/11 >65 y) and 9/11 (81.81%) of them had one or more comorbidities, corroborating the knowledge that advanced age and pre-existing medical conditions represent the major risk factors for developing a severe disease.
At the two time points, PBMCs were subjected to multiparametric flow cytometry analyses (Fig. S1A) and mapped by t-SNE plots (Fig. S2A). During the infection, COVID-19 patients, especially those with severe disease, experienced a reduction of T lymphocytes, particularly of CD8+ T cells, and a trend toward increased monocytes proportions, while the frequency of B lymphocytes was quite variable (Fig. 1A, B). On the contrary, natural killer (NK) cells were significantly expanded, especially in subjects with mild disease (Fig. 1A). The proportion of the different PBMC populations tended to normalize post-infection, except for a persistent increased frequency of NK cells.
PBMCs from healthy donors (HD, N=4), patients with mild symptoms during infection and post-infection (N=4), and severe disease during infection (N=7) and post-infection (N=6) phases analyzed by multiparametric flow cytometry. a) Frequency of monocytes, B lymphocytes, T lymphocytes, CD3+ CD56+ cells and NK cells is shown as percentage of live total PBMC. b) Frequency of CD4+ and CD8+ T lymphocytes is represented as percentage of live total PBMC. c) Relative abundance of CD8+ naïve, central memory (CM), effector memory (EM) and effector memory CD45RA+ (EMRA) cells shown as percentage of live total CD8+ T lymphocytes. d) Frequency of naïve B cells, total memory, non-switched memory, switched memory, memory IgM+, memory IgG+ and plasmablasts shown as percentage of live total B lymphocytes; for memory IgM+ and memory IgG+ from severe patients during infection and post-infection, N=5. e) IgM, IgG and IgA titers to SARS-CoV-2 nucleoprotein (N), receptor-binding domain (RBD), spike subunit 1 (S1) and subunit 2 (S2) measured by ELISA in the plasma of HD (N=5), mild patients during infection (N=4) and post-infection (N=4) and severe patients during infection (N=7) and post-infection (N=6). f) Neutralization of binding of recombinant RBD protein to a HEK293T cell line expressing hACE2 by sera of HD (N=4), mild patients during infection (N=4) and post-infection (N=4) and severe patients during infection (N=7) and post-infection (N=6). Positivity threshold: 50% of binding inhibition. a-f) Data are represented as box and whiskers showing median, min to max, and individual values. Statistical analyses were performed using Mann-Whitney t test to compare ranks. * p < 0.05; ** p < 0.01. In e) asterisk(s) above individual boxes denote statistical significance compared to HD, while specific comparisons are defined by square brackets colored according to the Ig isotype considered in the comparison.
” data-icon-position=”” data-hide-link-title=”0″>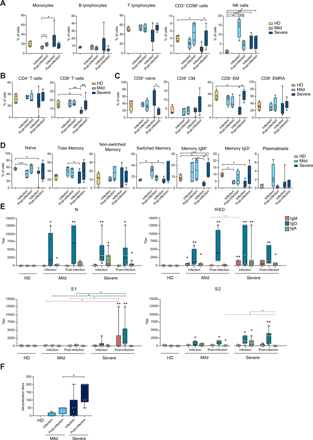

PBMCs from healthy donors (HD, N=4), patients with mild symptoms during infection and post-infection (N=4), and severe disease during infection (N=7) and post-infection (N=6) phases analyzed by multiparametric flow cytometry. a) Frequency of monocytes, B lymphocytes, T lymphocytes, CD3+ CD56+ cells and NK cells is shown as percentage of live total PBMC. b) Frequency of CD4+ and CD8+ T lymphocytes is represented as percentage of live total PBMC. c) Relative abundance of CD8+ naïve, central memory (CM), effector memory (EM) and effector memory CD45RA+ (EMRA) cells shown as percentage of live total CD8+ T lymphocytes. d) Frequency of naïve B cells, total memory, non-switched memory, switched memory, memory IgM+, memory IgG+ and plasmablasts shown as percentage of live total B lymphocytes; for memory IgM+ and memory IgG+ from severe patients during infection and post-infection, N=5. e) IgM, IgG and IgA titers to SARS-CoV-2 nucleoprotein (N), receptor-binding domain (RBD), spike subunit 1 (S1) and subunit 2 (S2) measured by ELISA in the plasma of HD (N=5), mild patients during infection (N=4) and post-infection (N=4) and severe patients during infection (N=7) and post-infection (N=6). f) Neutralization of binding of recombinant RBD protein to a HEK293T cell line expressing hACE2 by sera of HD (N=4), mild patients during infection (N=4) and post-infection (N=4) and severe patients during infection (N=7) and post-infection (N=6). Positivity threshold: 50% of binding inhibition. a-f) Data are represented as box and whiskers showing median, min to max, and individual values. Statistical analyses were performed using Mann-Whitney t test to compare ranks. * p < 0.05; ** p < 0.01. In e) asterisk(s) above individual boxes denote statistical significance compared to HD, while specific comparisons are defined by square brackets colored according to the Ig isotype considered in the comparison.
COVID-19 immune signatures
To identify specific immunological traits of patients with mild or severe disease, during and after the infection, we performed multiparametric FACS analyses of circulating T and B lymphocytes (Fig. S1B-E) and measured antibodies induced against the SARS-CoV-2 nucleocapsid (N) and spike (S) proteins in patients’ sera.
Among T lymphocytes, CD8+ cells from patients with severe disease showed a reduced frequency of effector memory cells (CD45RO+, CCR7-) and a decreased IFN-γ production capacity, during the infection (Fig. 1C, Fig. S1B, C and Fig. S2B), paralleled by an increased relative abundance of naïve cells. The same alterations were observed for CD4+ T cells, though less pronounced (Fig. S1B, D and Fig. S2B, C). The phenotyping of T helper cells indicated a moderate increase in the frequency of non-conventional TH1 (TH1*) cells in subjects with mild symptoms during the infection, that was reduced in patients with severe disease instead (Fig. S1D and Fig. S2D). After the resolution of the infection, all COVID-19 patients showed a significant impairment of the TH1 subset. In patients with severe disease, the frequency of TREG was moderately reduced during the infection and that of TH17 was increased post-infection, while the same subsets did not show any significant alteration in patients with mild disease (Fig. S1D and Fig. S2D).
Within the B cell population, total memory B cells were more abundant in subjects with mild disease during the infection. This difference was magnified when looking at switched memory B cells and specifically at IgM+ B lymphocytes, while the frequency of IgG+ B cells did not significantly differ between patients. However, the relative abundance of the switched memory B cells, and of the IgG+ ones in particular, was higher in severe patients post-infection. The frequency of plasmablasts was variable, with an increase that tended to be transient in mild patients and smaller but sustained in those with severe disease (Fig. 1D and Fig. S1E).
We measured the anti-SARS-CoV-2 antibody plasma levels by ELISA, assessing IgM, IgA and IgG polyclonal binding to the N protein, and to the N-terminal S1, the Receptor-Binding Domain (RBD), and the C-terminal S2 domains of the S protein. N and RBD elicited the highest antibody titers. RBD stimulated a rather homogeneous antibody response in all COVID-19 patients, while S1 and S2 tended to be better recognized by antibodies from subjects with a severe disease (Fig. 1E). Overall, anti-N and anti-RBD IgG were detected during the infection and had the highest and comparable titers in all patients’ groups. IgA were also detected against both proteins, and tended to be higher in severe patients. IgM were more abundant in patients with severe disease, mainly recognizing RBD, whereas anti-N IgM was almost undetectable (Fig. 1E).
To evaluate the presence of potentially protective antibodies, we tested the ability of plasma samples to block the binding of a recombinant RBD protein to a HEK293T cell line stably expressing the hACE2 receptor. Neutralization of binding was higher in severe patients compared to those with mild disease, and increased in both patient groups upon resolution of infection (Fig. 1F). Sera with neutralizing activity had detectable antibodies against S and RBD proteins, but we could not observe a clear correlation between anti-S antibody titers from a specific class and neutralization. Altogether, these data indicate a broad rearrangement of the adaptive immune system over time, involving both T and B lymphocytes, that was more evident in patients with severe disease.
Pervasive, graded and durable transcriptional changes in COVID-19 patients’ PBMC
To get deeper insights into the evolution of the immune response against SARS-CoV-2, we analyzed the transcriptional profile and the TCR and BCR repertoires at the single-cell resolution of PBMC from six COVID-19 patients, three mild and three severe, and two healthy controls. Four of the six COVID-19 patients, two mild and two severe, were profiled both during the infection (Day 1 – Day 16 from diagnosis) and about 3 weeks after the infection resolution (Day 19 – Day 21 from the negative swab, corresponding to Day 50 – Day 51 from diagnosis), enabling us to dissect the development of the anti-SARS-CoV-2 immunity over the course of the disease.
Clustering of total PBMCs scRNA-seq profiles identified five distinct populations corresponding to the main circulating immune cell types: monocytes, NK, T and B lymphocytes, and megakaryocytes (Fig. 2A), defined by the combined expression of selected lineage-specific genes (Fig. S3A). The disease severity deeply influenced the transcriptome of all populations, resulting in a graded segregation of HD from mild and severe COVID-19 patients during the infection (Fig. S3B, left panel). Such a pervasive effect was reduced post-infection, although the distribution of cells derived from patients was still clearly distinguishable from those of HD (Fig. S3B, right panel), indicating that the SARS-CoV-2 infection can affect the immunophenotype of exposed individuals for weeks after its resolution. Consistently with the literature (14–16), we observed a sizeable alteration of immune cells relative abundance in COVID-19 patients compared to HD both during the infection and post-infection (Fig. 2B). During the infection, T lymphocytes showed reduced frequencies in patients, especially in those with severe disease. Conversely, monocytes and megakaryocytes showed a progressive increase from HD to mild and severe COVID-19, while NK cells were especially expanded in patients with mild disease (Fig. 2B left panel). After resolution of the infection, we observed a general trend toward the normalization of immune population abundance in severe patients, except for a residual expansion of NK cells (Fig. 2B right panel). Mild patients retained an altered immune profile, with reduced T cell frequencies and an inflated innate immune compartment (monocytes, NK and megakaryocytes) (Fig. 2B right panel), suggesting a persistent inflammatory status.
Pervasive, graded and durable transcriptional changes in the immune populations in COVID-19 patients. Single-cell RNA-seq of PBMCs from 2 HD, 3 mild and 3 severe patients. a) Uniform manifold approximation and projection (UMAP) identified immune cell populations during infection (left) and post-infection (right). b) Barplots show the relative abundance of monocytes, NK cells, megakaryocytes, B lymphocytes and T lymphocytes identified during infection (left) and post-infection (right). Percentages represent the average value of the patient cohorts.
” data-icon-position=”” data-hide-link-title=”0″>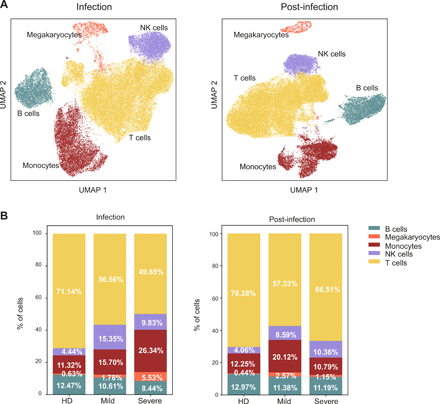

Pervasive, graded and durable transcriptional changes in the immune populations in COVID-19 patients. Single-cell RNA-seq of PBMCs from 2 HD, 3 mild and 3 severe patients. a) Uniform manifold approximation and projection (UMAP) identified immune cell populations during infection (left) and post-infection (right). b) Barplots show the relative abundance of monocytes, NK cells, megakaryocytes, B lymphocytes and T lymphocytes identified during infection (left) and post-infection (right). Percentages represent the average value of the patient cohorts.
Altogether, these transcriptomic data show that SARS-CoV-2 infection resulted in a long-lasting alteration of the circulating immune cell populations composition. This effect was particularly evident during the acute immune response, when immune cells are recruited to the infected tissues, but persisted after the infection resolved.
Elevated type I IFN signaling and reduced HLA-II expression in monocytes from COVID-19 patients
Innate immune cells contribute to the systemic inflammation that characterizes severe COVID-19 (5, 17). The appearance of monocytes with an altered immune profile has been described in COVID-19 patients, sometimes with contrasting features (18–20). Therefore, we investigated the phenotype of circulating monocytes in our patients’ cohort.
Transcriptional analysis identified seven monocytes clusters, one being largely populated by cells from HD (Mo 5) (Fig. 3A, B). During infection, monocytes from mild and severe patients were characterized by the prevalence of two clusters, Mo 1 and Mo 3, respectively (Fig. 3B and Table S2). Differential expression and gene ontology analyses showed that cluster Mo 1 expressed high levels of HLA-II genes, resembling monocytes differentiating into dendritic cells, while cluster Mo 3 was defined by the elevated expression of type I IFN responsive genes (Fig. 3C-E, Fig. S4A, B). Patients with severe disease were also characterized by the lack of non-classical monocytes (Mo 4), that have been associated with inflammation resolution (21), and which appeared after viral clearance (Fig. 3B). The post-infection phase was marked by the appearance of two additional clusters (Mo 6 and Mo 7), with cluster Mo 6 displaying activation (FOS, JUN, CD83) and pro-inflammatory (IL1B, CCL3, CCL4 and TNF) features, more expanded in mild patients (Fig. 3B, C).
Monocytes and NK cells phenotype in COVID-19 patients. a) Monocytes from HD and COVID-19 patients were segregated into 7 transcriptional clusters, visualized by UMAP. b) Barplot illustrates the relative abundance of the 7 subpopulations of monocytes in HD, mild and severe patients during infection and post-infection. Percentages shown are the average of the indicated cohort, and individual values are reported in Table S2. c) Heatmap of the top 10 differentially expressed genes in the 7 monocyte clusters. Violin plots show the expression of d) HLA-II genes and e) type I IFN-responsive genes in the indicated patient cohorts during and after infection. f) NK cells from HD and COVID-19 patients were divided into 3 transcriptional clusters, visualized by UMAP. g) Barplot illustrates the relative abundance of the 3 subpopulations of NK cells in HD and patients with mild and severe disease during infection and post-infection. Percentages shown are the average of the indicated cohort, and individual values are reported in Table S2. h, i) Dotplots showing the expression of the indicated subset-specific genes in HD and patients with mild and severe disease during infection and post-infection.
” data-icon-position=”” data-hide-link-title=”0″>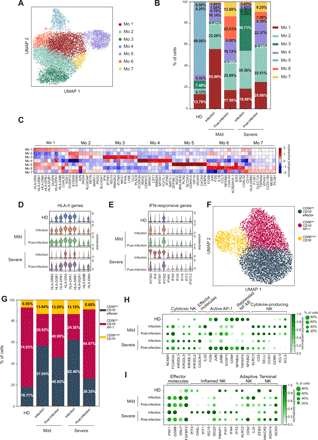

Monocytes and NK cells phenotype in COVID-19 patients. a) Monocytes from HD and COVID-19 patients were segregated into 7 transcriptional clusters, visualized by UMAP. b) Barplot illustrates the relative abundance of the 7 subpopulations of monocytes in HD, mild and severe patients during infection and post-infection. Percentages shown are the average of the indicated cohort, and individual values are reported in Table S2. c) Heatmap of the top 10 differentially expressed genes in the 7 monocyte clusters. Violin plots show the expression of d) HLA-II genes and e) type I IFN-responsive genes in the indicated patient cohorts during and after infection. f) NK cells from HD and COVID-19 patients were divided into 3 transcriptional clusters, visualized by UMAP. g) Barplot illustrates the relative abundance of the 3 subpopulations of NK cells in HD and patients with mild and severe disease during infection and post-infection. Percentages shown are the average of the indicated cohort, and individual values are reported in Table S2. h, i) Dotplots showing the expression of the indicated subset-specific genes in HD and patients with mild and severe disease during infection and post-infection.
These data indicate that monocytes from severe COVID-19 patients showed an up-regulated type I IFN response signature compared to patients with mild disease, and a considerable reduction of HLA-II genes expression (Fig. 3D, E), a proposed surrogate marker of immunoparalysis in sepsis (22). The impaired HLA-II genes signature may result from the decreased IFN-γ production in severe patients (Fig. S2B). Moreover, the appearance of a sub-population expressing pro-inflammatory genes post-infection may underlie the persistence of an inflammatory status.
Distinct activation of “adaptive” and “inflamed” transcriptional programs in NK cells from mild and severe COVID-19 patients
NK cells are crucial in the defense against viral infections (23). We observed a significant increase in the frequency of NK cells in patients with mild disease during the infection compared to the other experimental groups, both by flow cytometry (Fig. 1A) and scRNA-seq (Fig. 2B). Thus, we determined whether NK cells from patients with mild and severe disease also had distinct expression profiles. Transcriptional analysis identified 3 major NK cell clusters (Fig. 3F): clusters 0 and 1 were characterized by the low expression of NCAM1 (CD56) paralleled by a high expression of FCGR3A (CD16) and several KIRs (Fig. S4C), thus resembling CD56dim CD16+ cytotoxic NK cells (24). Clusters 0 and 1 showed limited differences, that were mostly confined to an elevated expression of the effector molecules CX3CR1 and IL32 in cluster 0 (hereafter CD56dim CD16+ effector), and the activation of AP-1 and repression of NF-κB pathways in cluster 1 (hereafter CD56dim CD16+ AP-1) (Fig. S4C). On the contrary, cluster 2 had high expression of NCAM1, KLRC1 (NKG2A), CD2, CD62L and CCR7 in the absence of FCGR3A and KIRs transcription (Fig. S4C), all features of CD56bright CD16– cytokine-producing NK cells which secrete abundant cytokines and proliferate in response to cytokine stimulation, but have limited cytotoxicity (24). The relative frequency of CD56dim CD16+ and CD56bright CD16– NK cells was increased in patients with mild disease during and post infection, and in patients with severe disease during the infection, while the proportion of the three NK subpopulations in individuals with severe disease post-infection was similar to the one observed in HD (Fig. 3G and Table S2).
Comparative transcriptional analysis showed that, during the infection, NK cells from mild patients expressed higher levels of genes typical of CD56bright CD16– cytokine-producing cells, such as KLRC1, GZMK, XCL1 and XCL2 (Fig. 3H). They also exhibited features of “adaptive” NK cells (25, 26), such as the expression of KLRC2, CD52 and IL32 (Fig. 3I). NK cells from severe patients up-regulated instead the transcription of genes characteristic of CD56dim CD16+ effector cells, like CX3CR1 and KIRs (Fig. 3H), but had an impaired expression of the activating receptor KLRC2 (NKG2C) and of some cytotoxic molecules, such as GNLY and FGFBP2 (Fig. 3I). NK cells from severe patients also expressed higher amounts of interferon responsive genes, characteristic of “inflamed” NK cells (25, 26) (Fig. 3I). Finally, NK cells post-infection, especially from patients with severe disease, up-regulated the expression of NCR3 (NKp30), HAVCR2 (TIM-3) and WDR74, that characterize terminally-differentiated NK cells (Fig. 3I). These data indicate that despite the similar subsets’ frequency, NK cells from patients with mild and severe disease activated distinct transcriptional programs which may underlie a different capacity to control the viral infection.
Increased frequency of memory B cells in COVID-19 patients.
Humoral immunity is key to neutralize viruses and to prevent reinfection. Thus, we explored the transcriptional phenotype of B lymphocytes to identify peculiar populations induced by SARS-CoV-2 infection. Clustering of gene expression profiles revealed five different B cell subpopulations (Fig. 4A) that were annotated based on the differential expression of selected markers (Fig. S5A) as: naïve (IGHD+ IGHM+), activated naïve (IGHD+ Nur77+), memory (CD27+), atypical memory (CD27+, CD21– and FCRL5+), and plasmablasts/plasmacells (MZB1+ CD38+). The relative proportion of the major B cell subsets from scRNAseq (Fig. 4B and Table S3) was similar to that measured by flow cytometry (Fig. 1D). During the infection, COVID-19 patients had an increased abundance of memory B lymphocytes, especially in subjects with mild disease (Fig. 4B). They were also characterized by the enrichment of a memory subset negative for CR2 (CD21) transcription and expressing high levels of FCRL5, resembling an atypical memory B cell population described in other infectious diseases (27, 28) (Fig. 4B and Fig. S5A). The identity of this population was confirmed by gene-set enrichment analyses (GSEA) (Fig. 4C and Table S4). These cells up-regulated CXCR3 and TBX21 (T-bet) that is required for IgG2a class switching (29, 30), a feature important for clearing viral infections (31). During the infection, B cells from severe patients were characterized by the up-regulation of several type I IFN-responsive genes, paralleled by a partial down-regulation of MHC-II genes (Fig. 4D), as seen in monocytes.
Features of B cell subsets, immunoglobulin classes, and clonal expansion in COVID-19 patients. a) UMAP depicting the 6 B cell clusters identified by transcriptional profiles. b) Barplot showing the relative abundance of each B cell cluster in HD, mild and severe COVID-19 patients during and after infection. c) GSEA showing the enrichment of the atypical memory B cells signature (Table S4) in the identified B cell subsets. d) Heatmap of type I IFN-responsive genes (top) and HLA-II genes (bottom) expression in B cells from HD, mild and severe patients during infection and post-infection. e) Immunoglobulin isotypes frequency in B cells from HD, mild and severe patients during and after infection. b, e) Percentages shown are the average of the indicated cohort, for individual values refer to Table S3. f) Donut plots representing B cell clonal expansion (outer circle) and isotype usage (inner circle) in HD, mild and severe patients during infection and post-infection. Each donut represents a single patient, and the number inside the donut is the number of cells analyzed.
” data-icon-position=”” data-hide-link-title=”0″>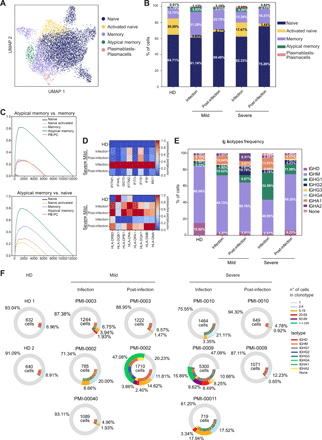

Features of B cell subsets, immunoglobulin classes, and clonal expansion in COVID-19 patients. a) UMAP depicting the 6 B cell clusters identified by transcriptional profiles. b) Barplot showing the relative abundance of each B cell cluster in HD, mild and severe COVID-19 patients during and after infection. c) GSEA showing the enrichment of the atypical memory B cells signature (Table S4) in the identified B cell subsets. d) Heatmap of type I IFN-responsive genes (top) and HLA-II genes (bottom) expression in B cells from HD, mild and severe patients during infection and post-infection. e) Immunoglobulin isotypes frequency in B cells from HD, mild and severe patients during and after infection. b, e) Percentages shown are the average of the indicated cohort, for individual values refer to Table S3. f) Donut plots representing B cell clonal expansion (outer circle) and isotype usage (inner circle) in HD, mild and severe patients during infection and post-infection. Each donut represents a single patient, and the number inside the donut is the number of cells analyzed.
Collectively, these analyses show an increased abundance of memory B lymphocytes in COVID-19 patients, especially in subjects with mild disease during the acute immune response, that were also characterized by the peculiar expansion of an atypical memory subpopulation. An elevated type I IFN response signature and the down-regulation of HLA-II genes expression were features of both monocytes and B cells from severe COVID-19 patients during the infection, possibly indicating an impaired antigen presentation capacity.
Ig isotypes and B cell clonal expansion during and post-infection
To evaluate B cell class switching and clonal expansion, we performed single-cell BCR sequencing analysis. We measured the proportion of IgA, IgD, IgG and IgM isotypes, while IgE was undetectable. IgM was the predominant immunoglobulin in all samples, while IgG and IgA isotypes were more abundant in COVID-19 patients compared to HD. In particular, patients with severe disease showed the highest levels of IgG1 and IgA1 isotypes during the infection (Fig. 4E and Table S3). The levels of IgG1 and IgA1 decreased post-infection, whereas IgG2 showed an opposite trend (Fig. 4E). Transcriptional profiles corroborated ELISA antibody measurement, revealing the preferential elicitation of IgA antibodies, especially against the RBD and N protein, in patients with severe disease (Fig. 1E).
Investigating the clonal expansion of circulating B cells, we observed a variegated response. During the infection, severe patients had a higher clonal expansion than mild patients, while post-infection we observed a generally reduced clonal expansion, with the exception of one of the two patients with mild disease (Fig. 4F). Expanded B cells included IgM, IgA2 and IgG subtypes. Notably, the B cell clones identified post-infection (Fig. S5B), likely-proliferating in response to SARS-CoV-2 antigens, did not match those captured during infection.
We also compared the preferential V(D)J gene usage in COVID-19 patients and HD. The IGHV3-23/IGHJ4 gene couple was enriched in all subjects, including HD, while the IGHV4-34/IGHJ6 pair was specifically overrepresented in severe patients during the infection. An enrichment of several gene segments (IGHV3-48/J4, IGHV3-49/J4, IGHV4-28/J4, IGHV4-34/J6, IGHV4-39/J5 and IGHV5-51/J6) was found in patients with mild disease post-infection, instead. The overrepresentation of IGHV4-34/IGHJ6 genes in patients may indicate a specific rearrangement induced by SARS-CoV-2. We observed a similar pattern for light chains with the enrichment of a single gene pair (IGKV1-9/IGKJ3) in severe patients during the infection, and of various combinations of gene segments in patients with mild disease post-infection (IGLV2-8/IGLJ2, IGLV2-14/IGLJ3, IGLV2-11/IGLJ1, IGKV1-5/IGKJ4, IGKV1D-33/IGKJ3, and IGKV1D-39/IGKJ2) (Fig. S5C, D). These results suggest a variable antibody response among COVID-19 patients, with increased frequencies of IgG2 and a broader Ig gene usage in patients with mild disease post-infection.
Altered composition of CD4+ and CD8+ T cell subsets in COVID-19 patients
Transient lymphopenia can characterize viral infections, can be induced by type I IFN response and can occur before the peak in the T cell response (32, 33). Although all patients had less T lymphocytes during the infection (Fig. 2B, left panel), subjects with mild disease showed a relative expansion of the CD8+ T cell population (Fig. 5A) indicating an ongoing CD8+-mediated immune response, possibly relevant for the management of SARS-CoV-2 infection. On the contrary, patients with severe disease experienced a relative contraction of the CD8+ lymphocytes compartment, resulting in an altered CD4/CD8 ratio (Fig. 5A).
Altered composition of CD4+ and CD8+ T cell subsets in COVID-19 patients. a) Barplot illustrates the percentages of CD4+ and CD8+ cells in CD3+ T lymphocytes of the patients analyzed. Percentages shown are the average of the cohort. b) UMAP representing the 11 T cell clusters identified by transcriptional analysis. c) Barplots describe the relative abundance of the identified CD4+ cell subsets in the cohorts analyzed during infection (left) and post-infection (right). d) Barplots show the relative abundance of the identified CD8+ lymphocytes subpopulations in the cohorts analyzed during infection (left) and post-infection (right). c-d) Percentages shown are the average of the indicated cohort, and individual values are reported in Table S5. e) Heatmap of type I IFN-responsive genes expression in the CD4+ T cell subsets. f) Frequency of CD8+ GZMB+, CD8+ GZMK+, and CD8+ GZMB+/GZMK+ cells detected by multiparametric flow cytometry in PBMCs of healthy donors (HD, N=4), mild patients during (N=4) and after (N=4) infection and severe patients during (N=7) and after (N=6) infection. Data represent the percentage of live total CD8+ T lymphocytes and are visualized as box and whiskers showing median, min to max and individual values. g) Dotplots showing the expression of CX3CR1 (top) and TCF7 (bottom) in the TC1 GZMK+ and TC1 GZMB+ clusters. h) GSEA analysis showing enrichment of the human effector T cell signature in CD8+ clusters. i) GSEA analyses showing enrichment of the human memory T cell signature (left) and human effector T cell signature (right) in the TC1 GZMK+ and TC1 GZMB+ clusters. Human effector and memory CD8+ T lymphocytes gene sets were derived from the analysis of the GSE100745 GEO Dataset (Table S7). j) GSEA analyses showing enrichment of terminal effectors KLRG1hi signature (left) and memory precursors KLRG1int signature (right) in the TC1 GZMK+ and TC1 GZMB+ clusters. Mouse KLRG1hi and KLRG1int gene sets were extracted from the Molecular Signature Database (GSE10239) (Table S7). In f) statistical analyses were performed using Mann-Whitney t test to compare ranks. * p < 0.05; ** p < 0.01.
” data-icon-position=”” data-hide-link-title=”0″>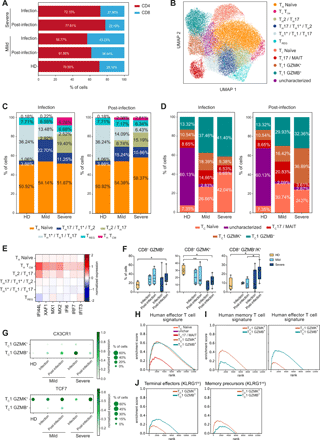

Altered composition of CD4+ and CD8+ T cell subsets in COVID-19 patients. a) Barplot illustrates the percentages of CD4+ and CD8+ cells in CD3+ T lymphocytes of the patients analyzed. Percentages shown are the average of the cohort. b) UMAP representing the 11 T cell clusters identified by transcriptional analysis. c) Barplots describe the relative abundance of the identified CD4+ cell subsets in the cohorts analyzed during infection (left) and post-infection (right). d) Barplots show the relative abundance of the identified CD8+ lymphocytes subpopulations in the cohorts analyzed during infection (left) and post-infection (right). c-d) Percentages shown are the average of the indicated cohort, and individual values are reported in Table S5. e) Heatmap of type I IFN-responsive genes expression in the CD4+ T cell subsets. f) Frequency of CD8+ GZMB+, CD8+ GZMK+, and CD8+ GZMB+/GZMK+ cells detected by multiparametric flow cytometry in PBMCs of healthy donors (HD, N=4), mild patients during (N=4) and after (N=4) infection and severe patients during (N=7) and after (N=6) infection. Data represent the percentage of live total CD8+ T lymphocytes and are visualized as box and whiskers showing median, min to max and individual values. g) Dotplots showing the expression of CX3CR1 (top) and TCF7 (bottom) in the TC1 GZMK+ and TC1 GZMB+ clusters. h) GSEA analysis showing enrichment of the human effector T cell signature in CD8+ clusters. i) GSEA analyses showing enrichment of the human memory T cell signature (left) and human effector T cell signature (right) in the TC1 GZMK+ and TC1 GZMB+ clusters. Human effector and memory CD8+ T lymphocytes gene sets were derived from the analysis of the GSE100745 GEO Dataset (Table S7). j) GSEA analyses showing enrichment of terminal effectors KLRG1hi signature (left) and memory precursors KLRG1int signature (right) in the TC1 GZMK+ and TC1 GZMB+ clusters. Mouse KLRG1hi and KLRG1int gene sets were extracted from the Molecular Signature Database (GSE10239) (Table S7). In f) statistical analyses were performed using Mann-Whitney t test to compare ranks. * p < 0.05; ** p < 0.01.
Transcriptional analysis of T lymphocytes identified 11 clusters, including 6 CD4+ and 5 CD8+ lymphocyte subsets (Fig. 5B), annotated based on inspection of selected marker genes (Fig. S6A). CD4+ T lymphocytes included: TH naïve, TH TCM, TH2/TH17, TH17/TH1*/TH2, TH1*/TH1/TH17 and TREG; while CD8+ lymphocytes were divided in TC naïve, TC1 GZMB+, TC1 GZMK+ and TC17/MAIT cells (Fig. 5B). CD8+ lymphocytes also contained a cluster that was difficult to characterize based on differentially expressed genes (dubbed as “uncharacterized”) but likely largely populated by naïve and central memory (TCM) cells from HD (Fig. 5B). The heterogeneous expression of critical marker genes suggests that these CD4+ lymphocyte clusters could not be simply catagorized as individual TH subsets. Instead, we observed a phenotypic gradient reflecting the transition from central memory (TH TCM and TH2/TH17) to effector memory subsets (TH17/TH1*/TH2 and TH1*/TH1/TH17), highlighted by the progressive reduction in the expression of key genes leading to the recirculation to secondary lymphoid organs (CCR7 and SELL) (34, 35) and regulating self-renewal capacity (TCF7 and LEF1) (36) (Fig. S6A).
While the proportion of TH naïve and TREG cells did not vary across patient cohorts, we observed an increase in central memory subsets (TCM and TH2/TH17) as disease severity progressed, especially during the infection (Fig. 5C and Table S5). In particular, severe patients were characterized by the skewing toward a TH2-like immune response, and by the appearance of a TCM cluster expressing the TFH marker CXCR5 and interferon responsive genes (Fig. 5C, E and Fig. S6A).
Within the CD8+ compartment, COVID-19 patients displayed a relative expansion of TC1 GZMB+ cells during the infection that increased with disease severity, while the frequency of TC1 GZMK+ lymphocytes was highest in severe patients post-infection (Fig. 5D and Table S5). These changes were confirmed at the protein level by flow cytometry where we observed a general increase in the frequency of GZMB+ cells in COVID-19 patients compared to HD and of GZMK+/GZMB+ double-positive cells in patients with severe disease post-infection (Fig. 5F and Fig. S1F). Both TC1 sub-populations expressed genes coding for effector molecules, such as CCL5, NKG7, PRF1, GZMA and CST7, though they were transcribed at higher levels in TC1 GZMB+ (Fig. S6A, C). The distinctive feature of the TC1 GZMB+ subset was an elevated production of GZMB, GNLY and FGFBP2 accompanied by the expression of CD16 (FCGR3A) and several killer cell lectin receptors, like KLRD1, KLRF1 and KLRC3, normally expressed on NK cells, indicating that these cells may be highly cytotoxic (Fig. S6B, left panel and Fig. S6C). Despite the expression of NK-related genes, TC1 GZMB+ cells displayed a variegated gene usage without a prevalence of the vα24-Jα18 genes in their TCR composition, indicating they were not invariant NKT (Fig. S6D and Table S6). TC1 GZMK+ cells were defined instead by the elevated expression of GZMK, CD160 and HLA-II genes (Fig. S6B, right panel and Fig. S6C). The elevated expression of HLA-DR and other HLA-II genes is associated with T cell activation and marks in vivo proliferating CD8+ T lymphocytes with reduced cytolytic activity (37), though granzyme K has been demonstrated to inhibit influenza virus replication (38). We explored the possibility that GZMB+ cells were short-lived effector lymphocytes (SLEC) while GZMK-expressing cells represented memory precursors effector cells (MPEC) (39, 40). Indeed, TC1 GZMB+ lymphocytes expressed elevated CX3CR1, characteristic of highly differentiated effector cells (41), while TC1 GZMK+ cells expressed TCF7 (Fig. 5G), a feature of memory-like cells that are able to proliferate in chronic viral infections (42). Moreover, TCF-1 (the TCF7 gene product) expression is a feature of SARS-CoV-2-specific memory T cells isolated from convalescent individuals (43). To further support the idea that TC1 GZMB+ lymphocytes are SLECs and TC1 GZMK+ lymphocytes are MPECs, we performed GSEA comparing the expression profile of the CD8+ subpopulations with the gene signatures of effector and memory human CD8+ T lymphocytes generated in response to vaccination with the live attenuated yellow fever virus (YFV) (44) (Table S7), one of the best-established models of acute viral infection in humans. Looking at the global transcriptional profile, both GZMB+ and GZMK+ cells showed a footprint of effector T lymphocytes (Fig. 5H), but focusing on the subset of differentially expressed genes the GZMK+ cells were enriched for a signature of memory lymphocytes (Fig. 5I). In addition, when compared to the gene-sets characterizing SLEC and MPEC defined by KLRG1 expression in a mouse model of LCMV acute infection (45) (Table S7), the GZMB+ cells showed a relative enrichment in the transcriptional profile of KLRG1hi terminal effectors, while GZMK+ cells in that of KLRG1int memory precursors (Fig. 5J).
Patients with mild disease had a higher frequency of TC17-like cells than severe patients (Fig. 5D). This subset was defined by the expression of KLRB1 (CD161), SLC4A10, RORC and CCR6 (Fig. S6A, C), and the TCR gene usage (TRAV1-2, TRAJ33/12/20 and TRBV20/6) demonstrated these were primarily composed of mucosal-associated invariant T (MAIT) cells (46, 47) (Fig. S6F and Table S6). Similar to TC1 GZMK+ cells, MAIT cells displayed a moderate expression of effector molecules-coding genes, such as CCL5, NKG7, PRF1, CST7 and, notably, GZMK (Fig. S6A-C), the latter being a feature of MAIT cells TCR-independent activation, resulting in a slower response with limited inflammation (48). MAIT cells showed an increased expression of the activation markers CD69, FOS and DUSP1 in patients with mild COVID-19 and the up-regulation of interferon responsive genes in patients with severe disease, during the infection (Fig. S6G).
Collectively, these data indicated a TCM– and TH2-skewed CD4+ T cell response in patients with severe disease accompanied by a type I IFN-responsive gene signature. CD8+ lymphocytes from COVID-19 patients were characterized by an elevated frequency of terminally differentiated GZMB+ effector cells during the infection, followed post-infection by an increased abundance of GZMK+ effector cells which may represent memory cell precursors.
CD4+ and CD8+ T cell clonal expansion in COVID-19 patients
To better dissect the SARS-CoV-2-specific T cell immune response, we speculated that expanded T cell clonotypes represented lymphocytes proliferating upon antigenic stimulation in response to SARS-CoV-2 infection, possibly accompanied by some bystander activation. To support the assumption, we generated antigen-specific, primary CD4+ T cell populations against SARS-CoV-2 RBD, S1 and N proteins and verified the appearance of expanded clonotypes identified by scTCR-seq analysis within these polyclonal populations. Indeed, we detected the TCRβ of 4 out of 6 clonotypes tested (Fig. 6A), providing evidence that a sizeable proportion of the expanded T lymphocytes identified by single-cell sequencing analyses are specific for SARS-CoV-2. Thus, we focused on the phenotypic characterization of the expanded T cell clones as a proxy for the SARS-CoV-2-specific T lymphocytes. We observed a higher clonal expansion for CD8+ T lymphocytes in COVID-19 patients compared to HD, although the absolute number of expanded cells was substantially lower in severe patients during the infection (Fig. 6B and Table S8). On the contrary, CD4+ T lymphocytes showed a limited clonal expansion even post-infection (Fig. 6C and Fig. S7A, B; Table S8). While the clonal expansion was distributed in all non-naïve CD8+ subsets, it was mostly confined to the TH1*/TH1/TH17 sub-population in TH cells (Fig. S7A, B and Table S9). However, the phenotype of expanded CD4+ T lymphocytes changed according to the disease severity: it was dominated by TH1*/TH1/TH17 and TH17/TH1*/TH2-like cells in mild patients, and by TH2/TH17 and TCM cells in patients with severe disease (Fig. 6D and Table S9). Also, the expanded CD4+ T lymphocytes showed an enhanced expression of effector molecules in mild patients, while those from patients with severe disease expressed higher amounts of pro-apoptotic genes, as well as SOCS1 and SOCS3 (Fig. S7C) that dampen the calcium signaling downstream the TCR (49) and inhibit STATs activation (50).
CD4+ and CD8+ T cell clonal expansion in COVID-19 patients. a) Primary CD4+ T cell populations specific to SARS-CoV-2 RBD, S1 and N proteins were generated from 2 patients with mild disease post-infection, and the presence of TCRβ chains of selected expanded clonotypes identified by scRNA-seq was detected by TCR-targeted SMART-qPCR assay. Barplots show CD8+ (b) and CD4+ (c) T cell clonal expansion in HD, mild and severe patients during and after infection. Numbers above the bars represent the total number of cells in the mentioned cohorts, and values inside the bars the percentage of cells in each group of clonal expansion; percentages represent the average of the cohort, and individual values are detailed in Table S8. d) Barplots show the relative abundance of the expanded CD4+ T lymphocytes in HD, mild and severe patients during and after infection. Percentages shown are the average of the indicated cohort, and individual values are reported in Table S9. Donut charts describe the clonal expansion in TC1 GZMB+ (e), TC1 GZMK+ (f), and TC17/MAIT (g) populations in the indicated patient cohorts and time points. Represented are: the total number of cells in the population inside the donut, and the mean percentage for each clonal expansion group; individual values for each patient are detailed in Table S9.
” data-icon-position=”” data-hide-link-title=”0″>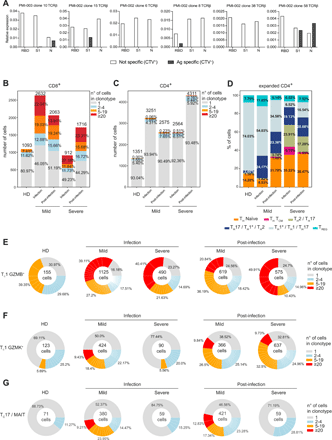

CD4+ and CD8+ T cell clonal expansion in COVID-19 patients. a) Primary CD4+ T cell populations specific to SARS-CoV-2 RBD, S1 and N proteins were generated from 2 patients with mild disease post-infection, and the presence of TCRβ chains of selected expanded clonotypes identified by scRNA-seq was detected by TCR-targeted SMART-qPCR assay. Barplots show CD8+ (b) and CD4+ (c) T cell clonal expansion in HD, mild and severe patients during and after infection. Numbers above the bars represent the total number of cells in the mentioned cohorts, and values inside the bars the percentage of cells in each group of clonal expansion; percentages represent the average of the cohort, and individual values are detailed in Table S8. d) Barplots show the relative abundance of the expanded CD4+ T lymphocytes in HD, mild and severe patients during and after infection. Percentages shown are the average of the indicated cohort, and individual values are reported in Table S9. Donut charts describe the clonal expansion in TC1 GZMB+ (e), TC1 GZMK+ (f), and TC17/MAIT (g) populations in the indicated patient cohorts and time points. Represented are: the total number of cells in the population inside the donut, and the mean percentage for each clonal expansion group; individual values for each patient are detailed in Table S9.
Among the CD8+ T cell subsets, TC1 GZMB+ cells were expanded in COVID-19 patients both during the infection and post-infection (Fig. 6E and Table S9), while TC1 GZMK+ lymphocytes showed an enhanced clonal expansion after the infection resolution (Fig. 6F and Table S9). Both TC1 GZMB+ and TC1 GZMK+ expanded clonotypes expressed high KLRG1 (Fig. S7D), a marker of effector T cells (51). In addition, expanded TC1 GZMB+ expressed TBX21 (T-bet), lost the expression of CD27 and CD28, and a small fraction of them up-regulated HAVCR2 (TIM-3) (Fig. S7D, E). Instead, expanded TC1 GZMK+ expressed CD27, whose interaction with CD70 on antigen-presenting cells (APC) promotes the generation and maintenance of memory cells (52), EOMES, TCF7, and very low CX3CR1 levels (Fig. S7D), resembling mouse effector T lymphocytes transitioning to memory precursors (53). Only a small fraction of expanded TC1 GZMB+ and TC1 GZMK+ cells expressed co-inhibitory receptors (Fig. S7E), suggesting they were not exhausted T lymphocytes. A small proportion of expanded TC1 GZMK+ cells also expressed CXCR5 and BCL6 (Fig. S7D), a feature of follicular cytotoxic CD8+ T lymphocytes, which can contribute to the control of chronic viral infections (54, 55).
Finally, Tc17/MAIT cells were exclusively expanded in patients with mild symptoms, both during the infection and post-infection (Fig. 6G and Table S9). The same trend of clonal proliferation was shown in the two patients that could be analyzed during the infection only (Fig. S7F). Notably, the patient with severe symptoms who completely lacked a CD8+ lymphocytes clonal expansion, suggesting he failed to mount an effective cytotoxic immune response, succumbed from the disease, similarly to what has been recently reported (13).
Altogether these data demonstrate that SARS-CoV-2 infection elicited a vigorous cytotoxic T cell immune response accompanied by a limited proliferation of T helper lymphocytes. The CD8+ response was dominated by the proliferation of GZMB+ cells throughout the infection, while GZMK+ cells were particularly expanded after the infection resolution, and possessed several transcriptional features associated to memory precursors.
Maintenance of highly expanded CD8+ clonotypes with focused plasticity within granzyme-producing subsets
To explore the clonal relationship between the different T cell subpopulations we monitored the evolution of the expanded clonotypes, grouped according to their phenotype, in each patient from the infection to the post-infection phase. Only the highly expanded clonotypes were retained throughout the course of the immune response (Fig. 7A, B and Fig. S8A, B; Table S10). Thus, only a minority of CD4+ lymphocytes were found both during the infection and post-infection, namely 37/211 clonotypes (17.54%) corresponding to 139/510 cells (27.25%). The few expanded clones were largely confined to the TH1*/TH1/TH17 subset (Fig. S8A, B) with limited plasticity toward phenotypically related populations, such as the TH17/TH1*/TH2 subset (Fig. S8C). Similarly, among the CD8+ T lymphocytes the most expanded clonotypes were those retained after the resolution of infection, but their higher clonality resulted in the sharing of 118/162 clonotypes (72.84%), corresponding to 2213/2533 cells (87.34%). To substantiate this finding, we analyzed CD8+ T lymphocytes isolated 10 months after the infection from two of the patients with mild disease. Only a small fraction (12.55% and 16.64%) of the clonotypes not expanded during the infection was detected at this time point, while most of the expanded clonotypes persisted, with an increased frequency of lymphocytes derived from the TC1 GZMK+ subset (Fig. S8D). The majority of proliferating clones during and post-infection were TC1 GZMB+ cells, but they showed a relatively high plasticity toward the TC1 GZMK+ subset (Fig. 7C), supporting the idea that the two populations may represent different developmental stages in the immune response rather that two functionally distinct clusters. Instead, MAIT cells constituted a clonally-independent CD8+ subpopulation (Fig. 7C), as expected due to their restricted TCR gene usage.
Heatmaps demonstrate clonotype sharing between CD8+ T cell clusters from infection (I) to post-infection (P-i) phases in mild (a) and severe (b) COVID-19 patients. TCR-α (TRA) and TCR-β (TRB) CDR3 sequences are listed in Table S10. c) STARTRAC transition index describes the degree of clonotype sharing and plasticity among the different CD8+ T cell clusters in the two mild and two severe patients analyzed. d) RNA velocity vectors plotted on a UMAP representing CD8+ T lymphocytes from COVID-19 patients. The yellow box highlights a subset of TC1 GZMB+ cells predicted to transdifferentiate into TC1 GZMK+ cells.
” data-icon-position=”” data-hide-link-title=”0″>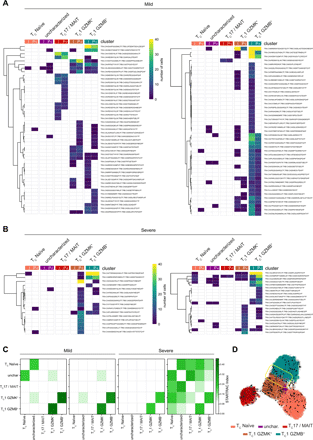

Heatmaps demonstrate clonotype sharing between CD8+ T cell clusters from infection (I) to post-infection (P-i) phases in mild (a) and severe (b) COVID-19 patients. TCR-α (TRA) and TCR-β (TRB) CDR3 sequences are listed in Table S10. c) STARTRAC transition index describes the degree of clonotype sharing and plasticity among the different CD8+ T cell clusters in the two mild and two severe patients analyzed. d) RNA velocity vectors plotted on a UMAP representing CD8+ T lymphocytes from COVID-19 patients. The yellow box highlights a subset of TC1 GZMB+ cells predicted to transdifferentiate into TC1 GZMK+ cells.
To highlight the relationship between the two granzyme-producing subsets we performed RNA velocity analysis (56). CD8+ T lymphocytes from COVID-19 patients showed an increased length of the velocity vectors compared to those from HD (Fig. S9A), indicating an ongoing alteration of the cell state. The RNA velocity was higher for the effector subsets and reduced post-infection (Fig. S9A), suggesting a trend toward a partial restoration of quiescence following the pathogen clearance. CD4+ T lymphocytes showed a similar picture (Fig. S9B). A subset of TC1 GZMB+ cells was predicted to transdifferentiate into TC1 GZMK+ cells (Fig. 7D) further supporting a developmental trajectory where a fraction of short-lived effectors fuels the generation of memory precursors.
To reveal whether evolutive pressures acted on the selection of expanded clonotypes preserved after infection resolution, we compared the complementarity-determining regions (CDR1, CDR2 and CDR3) aminoacidic sequences of expanded clonotypes during the infection and post-infection. Although we identified only a “public” clonotype among two mild patients, defined as a complete CDR1, CDR2 and CDR3 homology, we observed a small enrichment of a limited set of CDR1 and CDR2 in the TCR of expanded lymphocytes post-infection (Fig. 8).
Sharing of CDR1 (left panels) and CDR2 (right panels) from the expanded clonotypes of 4 COVID-19 patients, two mild and two severe, during infection (top panels) and after infection (bottom panels). The percentage of shared regions (s) is shown. Overlapping clonotypes are colored as indicated in the heatmap. In each panel, the inset is a zoom-in of the area highlighted by the grey rectangle.
” data-icon-position=”” data-hide-link-title=”0″>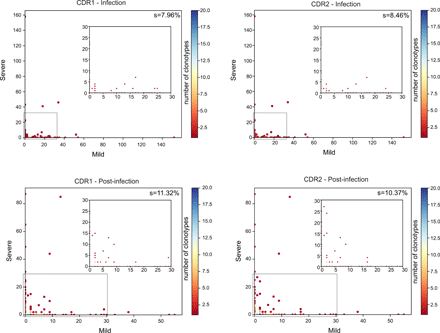

Sharing of CDR1 (left panels) and CDR2 (right panels) from the expanded clonotypes of 4 COVID-19 patients, two mild and two severe, during infection (top panels) and after infection (bottom panels). The percentage of shared regions (s) is shown. Overlapping clonotypes are colored as indicated in the heatmap. In each panel, the inset is a zoom-in of the area highlighted by the grey rectangle.
Collectively, these data demonstrated that only highly proliferating clonotypes were maintained after resolution of infection, part of which probably forming a pool of SARS-CoV-2-specific memory cells. A proportion of the CD8+ expanded clonotypes were found both in the TC1 GZMB+ and in the TC1 GZMK+ subsets indicating a common ancestry. Finally, TCR composition analysis of the expanded clonotypes retained post-infection revealed a diversity in their repertoire but also the selection of a small set of common CDRs, suggesting the presence of some physical constraint needed for the recognition of cognate antigens.
DISCUSSION
Knowledge on the temporal evolution of the immune response in COVID-19 patients is especially valuable in interpreting SARS-CoV-2 adaptive immunity and for the optimization of effective vaccines. In this study, we integrated single-cell phenotypic and repertoire analyses to investigate the immune response in COVID-19 patients with mild and severe symptoms during the acute disease and after the resolution of the infection. The collected data describe the development of immune responses in these patients, suggesting a prominent role for CD8+ T lymphocytes in clearing SARS-CoV-2 infection.
We confirmed that SARS-CoV-2 infection results in a profound remodelling of the circulating immune cell populations, especially in patients with a severe disease (12, 57, 58). The immune remodeling is echoed by pervasive, graded and durable changes at the transcriptional level (15, 17) that lasted weeks after the infection resolution. A deeper understanding of this long-lasting perturbation may provide new insight on the post-COVID-19 syndrome (59). We observed that the extensive transcriptional alteration was characterized by a pervasive type I IFN-response signature that crossed all the immune populations, especially involving monocytes. Type I IFN signaling has a prominent role in the innate immune response against viruses and its impairment or temporal dysregulation associates with severe COVID-19 (11, 60, 61). Some studies highlight that type I IFN signaling is lower in COVID-19 compared to other respiratory viral infections (19, 62). Although the intensity of the interferon response is higher in subjects with severe disease compared to those with mild symptoms (18) and correlates with the viral load (62), but the opposite scenario is observed in critically ill patients (63). We identified an up-regulated type I IFN-responsive gene signature in patients with severe disease compared to individuals with mild symptoms, paralleled by an impaired antigen presentation transcriptional program and the lack of non-classical monocytes, other prominent features of severe COVID-19 patients (5, 20, 64, 65).
NK cells are crucial in the defense against viral infections, as they kill infected cells and bridge the gap between the innate immune response and the setting of an optimal adaptive immunity. We found an increased frequency of NK cells in patients with mild disease during the infection, including both CD56dim CD16+ effector, CD56bright CD16– cytokine-producing and “adaptive” NK cells, which have been originally identified in human cytomegalovirus infections and can form a pool of memory-like cells (24). On the contrary, NK cells from patients with severe disease showed an impaired production of cytotoxic molecules during the infection and were skewed toward an “inflamed” phenotype, whose impact on their antiviral activity will require additional investigation.
Pathogen-specific antibodies are fundamental to provide protection against virus reinfection. The magnitude of antibody response correlates with viral load and disease severity (66). Coherently, our data showed that the amount of neutralizing antibodies increased with disease severity and was higher post-infection. Though, the neutralization titers in severe patients did not perfectly match the kinetics of the antibody response against RBD, suggesting that antibodies directed to other S domains may contribute to neutralize the binding of RBD to the host cells. Our data confirmed that the N and RBD proteins represent immunodominant antigens eliciting the highest IgG titers (13, 67, 68). Although the antibody response was dominated by IgG, we also detected IgA primarily in patients with severe disease, which could contribute to the tissue damage in severe COVID-19 (69). The IgM response was also higher in patients with severe disease, mostly directed against the RBD, and persisted post-infection, suggesting that these patients might have experienced a delayed elicitation of the antibody response.
Along with an overall increase of memory B cells in COVID-19 patients, we found the expansion of an atypical memory subpopulation during the infection (70, 71). Atypical memory B lymphocytes are thought to be functionally impaired but recent evidence suggests they are instead mature and optimally responsive cells (72, 73). Also, their elevated T-bet expression may underlie an activation state and suggest they can mount a T cell-independent immune response (30, 74). This feature, together with the ability to induce IgG2a class switching (75), may contribute to improved antiviral protection since viruses can act as T cell-independent antigens in vivo (76).
As for monocytes, we observed in B cells the up-regulation of various type I IFN-responsive genes, specifically in patients with severe disease during the infection (14, 19, 20). Moreover, the down-regulation of HLA-II genes expression in these cells indicated that SARS-CoV-2 infection can impair the antigen presentation capacity of different kind of APCs. Finally, analysis of B cell repertoire revealed that expanded B cell clones detected during the infection were distinct from those retrieved post-infection. A prolonged patients’ monitoring would be needed to correlate the presence of expanded clonotypes with the acquisition of humoral immunity.
T lymphocytes are pivotal in tackling viral infections and establishing a protective immunological memory, as cytotoxic CD8+ T lymphocytes kill infected cells and CD4+ T helper cells provide the signals to optimize effective and durable adaptive immune responses (77, 78). T cells underwent an extensive remodeling in COVID-19 patients both in terms of abundance and phenotype. We found that T helper lymphocytes from subjects with mild disease were enriched in TH17- and TH1*-like cells. T helper lymphocytes were partially skewed, instead, toward a TCM and TH2-like phenotype in patients with severe disease. The same trend was maintained and even enhanced when focusing on the expanded clonotypes, that were enriched in SARS-CoV-2-specific T cells. Interestingly, an enhanced type 2 immune response is seen in fatal SARS-CoV infections (79) and severe COVID-19 cases (11, 80), and has detrimental effects in other respiratory infections (81). During the infection, CD4+ T lymphocytes from severe patients also displayed the appearance of a TCM-like subset characterized by a type I IFN-response signature. Among the genes up-regulated in this signature, the X-linked inhibitor of apoptosis (XIAP)-associated factor 1 gene (XAF1) triggers apoptosis under stress conditions (82) and can induce increased T lymphocyte apoptosis in COVID-19 patients (18), suggesting that an impaired T cell survival may represent one of the factors leading to lymphopenia in patients with severe disease. In addition, we found that expanded CD4+ T lymphocytes from patients with severe disease expressed higher amounts of pro-apoptotic genes and lower levels of effector molecules, indicating an impaired fitness and a possible defect in mounting an effective antiviral response.
CD8+ T lymphocytes have the largest contraction in absolute numbers in COVID-19 patients during infection (12, 15, 83). Despite this, we observed a relative increase in the frequency of non-naïve T lymphocytes in patients compared to HD. These cells could be distinguished in granzyme B+, granzyme K+ and MAIT cells. The different proportions of CD8+ T lymphocytes in mild and severe COVID-19 patients may suggest qualitative or temporal differences in the cell-mediated immune response depending on disease severity.
The longitudinal integrated analyses of transcriptomes and TCR repertoires in COVID-19 patients coupled with immunophenotyping allowed us to depict the evolution of T cell responses in SARS-CoV-2-infected individuals by comparing and contrasting the phenotype of clonally expanded T lymphocytes as a proxy for antigen-specific cells. Here, we show that the expanded clonotypes detected by scTCR-seq included some SARS-CoV-2-specific T cells. We observed an enhanced CD8+ T lymphocyte clonal expansion in COVID-19 patients compared to HD, indicating an ongoing adaptive cellular response in patients, although it was curbed in subjects with severe disease. The CD8+ T cell clonal expansion was paralleled by less evident CD4+ T lymphocytes clonal expansion, similarly to what is described in other acute viral infections (84). Among CD8+ T lymphocytes we found that GZMB+ effector cells, expressing high levels of cytotoxic molecules, were considerably expanded during the infection and post-infection in all COVID-19 patients, regardless of disease severity. On the contrary, GZMK+ lymphocytes, which showed several features of memory-like cells and had lower expression of effector molecules, were preferentially expanded post-infection and were retained in higher proportions months after the clearance of the pathogen, likely representing MPECs. The appearance of MPECs may be indicative of a resolution of the viral infection, since a curtailed antigenic stimulation during the later stages of infection enhances the generation of memory cells, while the continuous exposure to antigenic stimuli drives the differentiation of terminal effectors (45). Thus, the appearance of expanded TC1 GZMK+ lymphocytes might represent a prognostic factor of COVID-19 resolution. Another interesting observation was the presence of expanded MAIT cells specifically in mild patients. Although these cells are primarily characterized for their capacity to specifically recognize bacterial and fungal metabolites presented by the MHC-class I-like molecule MR1 (85), they have antiviral capacity in response to bystander activation (86). Whether MAIT cells can restrain SARS-CoV-2 replication and which factors modulate their frequencies in COVID-19 patients requires further investigation.
In conclusion, our data demonstrated that activation of cytotoxic T lymphocytes marked natural immune response against SARS-CoV-2. In line with the evidence that coordinated T and B cell immune responses positively correlated with COVID-19 prognosis (13), a variety of effective vaccines against SARS-CoV-2 have been developed that elicit antigen-specific CD4+ and CD8+ T lymphocytes and SARS-CoV-2 neutralizing antibodies. The extent of the duration of the protective immunity to SARS-CoV-2 induced by infection and vaccines is a subject of ongoing discussion. SARS-CoV-specific memory T lymphocytes last many years after infection (9, 87), and SARS-CoV-2 specific memory T and B cells persist for months after the infection (88–91), hinting at long-term immunity. The detection of antigen-specific MPECs may represent an early-stage correlate of durable memory. The limited clonal expansion we observed for CD4+ T lymphocytes was in contrast to that of CD8+ T lymphocytes, but in line with that observed in virus-naïve individuals in other models of acute viral infections, like the YFV vaccination. YFV recall vaccination induces higher expansion of antigen-specific CD4+ T cells compared to the first challenge (84). It will be interesting to verify whether anti-SARS-CoV-2 vaccines will be able to boost a similar T helper clonal expansion to elicit a long-lasting protective immunity, and to compare the immunological memory induced by natural infections and vaccines.
Collectively, this study describes the development of an adaptive immune response in individuals who had mild or severe COVID-19. It provides insights into the generation of a T cell-driven immune response to SARS-CoV-2, by delineating the features of an effector GZMK+ CD8+ T lymphocyte population which may generate protective memory against SARS-CoV-2.
MATERIALS AND METHODS
More details on all of these techniques can be found in the supplemental material.
Study Design
The objective of the study was to describe the immune response to SARS-CoV-2 infection in patients with different grades of COVID-19 disease severity. Serological, phenotypic and transcriptomic analyses were combined to describe features associated with SARS-CoV-2 infection. We investigated innate and adaptive immune responses in 17 patients, with mild (N=6) or severe (N=11) disease, during and after infection, and compared to healthy individuals (N=5).
Sample collection. Patients enrolled in the study were diagnosed based on a RT-PCR positive nasopharyngeal swab for SARS-CoV-2. They were classified as affected by severe disease (radiological diagnosis of pneumonia and/or respiratory failure, i.e., pO2 <80 mmHg or pCO2 <35 mmHg or satO2 37.5°C, cough or coryza, without evidence of pneumonia and/or respiratory failure). The study was approved by the Institutional Review Board Milano Area 2 (#331_2020).
PBMC isolation. In brief, PBMC were isolated by density-gradient centrifugation following the Ficoll-Paque Plus standard protocol.
FACS immunophenotyping. In brief, 100,000 cells were washed in PBS 1X, stained with Fixable Viability Stain 15’ RT, and then washed in MACS buffer. For the detection of cell-surface proteins, cells were stained with the indicated antibodies diluted in Brilliant Stain Buffer solution (1:2 in MACS buffer) for 30’ RT. After washing in MACS buffer, cells were fixed 15’ at 4°C using the eBioscience FOXP3 staining kit and washed again in MACS buffer prior to acquisition. For intracellular cytokine staining, cells were stimulated for 4 hours with 0.2μM PMA and 1μg/ml ionomycin in culture medium at 37°C. 10μg/ml BFA was added in the last 2h of stimulation. To detect intracellular proteins, cells were permeabilized and stained in Permeabilization Reagent supplemented with antibodies for 30’ at 4°C. All incubation steps were performed in the dark. Antibodies used for FACS are listed in Table S11.
Expression and purification of SARS-CoV-2 proteins. In brief, based on an early SARS-CoV-2 sequence isolate (Wuhan-Hu-1), human codon-optimized nucleotide sequences encoding the subunits of the spike glycoprotein, S1 (aa 2-673), S2 (aa 686-1211), and RBD (aa 318-541), and the full-length nucleocapside protein, were cloned into a pcDNA 3.4 vector. Recombinant proteins included a custom N-terminal signal peptide for protein secretion and a C-terminal octa-histidine tag for purification. Plasmids were transiently transfected into Expi293TM cells and after 72h recombinant proteins were purified from culture supernatants by IMAC.
ELISA . Anti-S1, -S2, -RBD and -N IgG, IgM and IgA plasma titers were determined by ELISA. In brief, 96-well plates were coated overnight at 4°C with 250ng/well of each recombinant protein in PBS 1x. After blocking with PBS/BSA 5%, plasma samples were serially diluted and incubated for 1 hour at 37°C. Plates were washed with PBS/Tween 0.5% and probed with HRP-conjugated α-human IgG, IgM and IgA secondary antibodies (1:1000 in PBS/BSA 1%/Tween 0.5%) for 40’ RT. After washing the reaction was developed with TMB for 10’, stopped with 100μl 1M H2SO4 and the absorbance measured at λ = 450 nm.
Neutralization of binding assay. In brief, to evaluate the concentration of serum neutralizing antibodies, 10μl of purified recombinant RBD-AlexaFluor647 (10μg/ml in PBS/FCS 1%) were mixed with 10μl of different sera dilutions in U bottom 96-well plates for 1h RT. After incubation, 30×103 HEK293TN-hACE2 cells were resuspended in 5μl PBS/FCS 1%, added to the mix and incubated 1h at 4°C. Unbound protein was removed by washing with PBS and RBD binding was detected by flow cytometry. Specific neutralization was calculated as follows: NOB (%) = 1–(MFISample– MFIbackground)/(MFICtrlNegative – MFIbackground).
Generation of hACE2 cell line. In brief, a cell line stably expressing hACE2 receptor (HEK293TN-hACE2) was generated by lentiviral transduction of HEK293TN cells with pLENTI_hACE2_HygR, obtained by hACE2 sub-cloning from pcDNA3.1-hACE2 into pLenti-CMV-GFP-Hygro, and now available to the scientific community through Addgene (#155296). Lentiviral particles were produced by calcium phosphate-based cotrasfection of 3rd generation helper and transfer plasmids, following standard procedures. 48h after transduction, HEK293TN were subjected to hygromycin selection. Expression of hACE2 was confirmed by flow cytometry.
Generation of antigen-specific primary CD4+ T cells. Briefly, to generate SARS-CoV-2-specific polyclonal T cell populations, monocytes from patients’ total PBMCs were isolated with human CD14 microbeads, irradiated (45Gy) and loaded with the recombinant S1, RBD and N proteins, by incubating 1×105 cells with 4μg/ml of antigen in complete medium for 6h. Loaded monocytes were then co-incubated with sorted and CTV-stained total memory CD4+ T cells at 1:2 ratio in complete medium in flat bottom 96-well plates. On day 3, cells were resuspended and transferred to U bottom 96-well plates. On day 6, CTV– were sorted and expanded with allogeneic irradiated feeder cells and phytohemagglutinin (1μg/ml) in complete medium containing IL-2 (500U/ml).
TCR-targeted SMART-qPCR. Briefly, to identify the presence of individual T cell clonotypes in the polyclonal SARS-CoV-2-specific T cell populations a method was set up based on a modified SMART-seq2 protocol.
scRNA-seq. PBMCs were thawed in culture medium. Viability was measured immediately before chip loading, exceeding 75% in all samples. 10,000 PBMC/sample were loaded on a Chromium 10X Controller to generate single-cell GEMs, according to Chromium Next GEM Single Cell 5′ Library & Gel Bead Kit v1.1 protocol. Gene expression, TCR and BCR enriched libraries were produced using the Chromium Single Cell 5′ Library Construction Kit according to manufacturer instructions. Indexed libraries were sequenced on an Illumina Novaseq Flow-Cell Type S2 (2x 150bp paired-end).
scRNA-seq data processing and quality control. Raw fastQ files were aligned against the GRCh38 human reference genome and quantified using Cell Ranger Single-Cell 10X pipeline with default parameters. The filtered cell barcode matrices of each sample obtained by Cell Ranger count were processed using Scanpy (v1.4.2). Samples were grouped based on the stage of the disease (Infection vs. Post-infection). To identify cell populations, we performed separated analyses on the two stages. The different batches were aggregated with concatenate function with basic QC filtering. For each sample, cells that expressed <200 genes and genes detected in less than 0.1% of the total cells were filtered out. A second filter was applied to remove low quality cells based on the median absolute deviation obtained from the distribution of: the number of expressed genes per cell, UMI counts and the percentage of mitochondrial and riboprotein genes. The obtained dataset was then normalized and log-transformed using Scanpy pp.normalize_per_cell and pp.log1p functions, and technical sources of variation were regressed out by pp.regress_out. For the subsequent analyses a set of high variable genes was identified with pp.highly_variable_genes (mean expression ranging from 0.0125 and 5 and dispersion greater than 0.01).
Dimensionality reduction and clustering. Briefly, PCA (tl.pca function) was used to identify significant principal components in the dataset. Selection of the number of components for the nearest neighbors network computation (pp.neighbors) was based on their visualization in an elbow plot (pl.pca_variance_ratio). Uniform manifold approximation and projection was performed for the spatial visualization of the single cell dataset and features. Finally, cells were clustered using the Leiden algorithm.
Differential expression analyses. Differentially expressed genes between the distinct clusters and experimental groups were identified with the tl.rank_genes_groups function and filtered based on adjusted P value (≤0.05) and log2 fold change (≥1). Gene Ontology analyses on differentially expressed genes were performed using Metascape with GO Biological Processes as Pathway and default parameters.
RNA velocity. RNA velocity analysis was performed using the scvelo python package (v.0.2.3), applying a dynamical model. The number of spliced and unspliced reads was counted directly on the Cell Ranger output and the calculated RNA velocity vectors were embedded in a UMAP space. Velocities were estimated by inferring the splicing kinetics of the top 50 differentially expressed genes in each population.
TCR-seq and BCR-seq. Raw fastQ files were assembled using the Cell Ranger VDJ 10X pipeline with default parameters to obtain CDR3 sequences and the rearrangement of V(D)J genes. For TCRs, only cells with a productive TCR alpha and beta chains pair and for BCRs only those with at least a productive heavy and light chains pair were considered for further analyses. A clonotype was defined by consistent CDR3 amino acid sequence, and V and J genes usage. Cells sharing the same clonotype were considered clonal, and the clonotype as clonally expanded.
STARTRAC transition index. For clonally expanded CD4+ (clonotypes ≥2 cells) and CD8+ (clonotypes ≥5 cells) T cells, STARTRAC Transition Index for each batch was calculated as described (92). Plot and figures were generated using seaborn 0.10.1 and matplotlib 3.3.1.
Statistics. Statistical analyses of flow cytometry and serological data were performed with GraphPad Prism software. Statistical significance between groups was determined using Mann-Whitney test to compare ranks. p values ≤ 0.05 were considered significant.


