Defenses against SARS-CoV-2 variants
Our key defense against the COVID-19 pandemic is neutralizing antibodies against the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus elicited by natural infection or vaccination. Recent emerging viral variants have raised concern because of their potential to escape antibody neutralization. Wang et al. identified four antibodies from early-outbreak convalescent donors that are potent against 23 variants, including variants of concern, and characterized their binding to the spike protein of SARS-CoV-2. Yuan et al. examined the impact of emerging mutations in the receptor-binding domain of the spike protein on binding to the host receptor ACE2 and to a range of antibodies. These studies may be helpful for developing more broadly effective vaccines and therapeutic antibodies.
Science, abh1766, this issue p. eabh1766, abh1139, this issue p. 818
Abstract
Neutralizing antibodies (nAbs) elicited against the receptor binding site (RBS) of the spike protein of wild-type severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) are generally less effective against recent variants of concern. RBS residues Glu484, Lys417, and Asn501 are mutated in variants first described in South Africa (B.1.351) and Brazil (P.1). We analyzed their effects on angiotensin-converting enzyme 2 binding, as well as the effects of two of these mutations (K417N and E484K) on nAbs isolated from COVID-19 patients. Binding and neutralization of the two most frequently elicited antibody families (IGHV3-53/3-66 and IGHV1-2), which can both bind the RBS in alternative binding modes, are abrogated by K417N, E484K, or both. These effects can be structurally explained by their extensive interactions with RBS nAbs. However, nAbs to the more conserved, cross-neutralizing CR3022 and S309 sites were largely unaffected. The results have implications for next-generation vaccines and antibody therapies.
The COVID-19 pandemic has already lasted for more than a year, but new infections are still escalating throughout the world. Although several different COVID-19 vaccines have been deployed globally, a major concern is the emergence of antigenically distinct severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants of concern (VOCs). In particular, the B.1.1.7 lineage that arose in the UK (1) and quickly became dominant, the B.1.351 (also known as 501Y.V2) lineage in South Africa (2), the B.1.1.28 lineage (and its descendant B.1.1.28.1, also known as P.1/501Y.V3) in Brazil (3), and B.1.232/B.1.427/B.1.429 (also called CAL.20C and CAL.20A) in the United States (4) have raised serious questions about the nature, extent, and consequences of antigenic drift in SARS-CoV-2. In the receptor binding site (RBS) of the spike (S) protein receptor-binding domain (RBD), the B.1.1.7 lineage has acquired an Asn501 → Tyr (N501Y) mutation, the B.1.351 and P.1 lineages share this mutation along with Lys417 → Asn/Thr (K417N/T) and Glu484 → Lys (E484K), whereas the California variants have a Leu452 → Arg mutation that is also present in the B.1.617 variant, which was first isolated in India, with Glu484 → Gln (5). E484K has also been detected in a few B.1.1.7 genomes (1) (Fig. 1A). We therefore investigated the structural and functional consequences of such mutations on neutralizing antibodies (nAbs) isolated from COVID-19 convalescent patients, as well as their effect on angiotensin-converting enzyme 2 (ACE2) receptor binding.
(A) Emergent mutations (spheres) in the RBS of B.1.351 and P.1 lineages are mapped onto a structure of SARS-CoV-2 RBD (white) in complex with ACE2 (green) (PDB 6M0J) (90). Binding affinities of Fc-tagged human ACE2 against SARS-CoV-2 RBD wild type and mutants were assayed by biolayer interferometry (BLI) experiments. Detailed sensorgrams are shown in fig. S1. (B) Distribution of IGHV gene usage. Numbers of RBD-targeting antibodies encoded by each IGHV gene are shown. The frequently used IGHV3-53 and IGHV3-66 genes are highlighted in blue, and IGHV1-2 in orange. The IGHV gene usage in 1593 SARS-CoV-2 RBD-targeting antibodies (11, 14–44) relative to healthy individuals (baseline) (76) (fold enrichment) is shown as black line segments. Hashtags (#) denote IGHV gene frequencies in healthy individuals that were not reported in (76). Asterisks denote IGHV genes that are significantly enriched over the baseline repertoire (76) (P < 0.05, one-sample proportion test with Bonferroni correction). A fold enrichment of 1 (red dashed line) represents no difference over baseline. (C) Effects of single mutations on the neutralization activity and binding affinity of each neutralizing antibody. “–” denotes an increase in half-maximal inhibitory concentration (IC50) or KD by a factor of 100. Results in red with “×” indicate that no neutralization activity or binding was detected at the highest amount of immunoglobulin G used. N.C., not categorized in the original studies; N.S., no structure available. (D) Neutralization of pseudotyped SARS-CoV-2 virus and variants carrying K417N or E484K mutations. We tested a panel of 17 neutralizing antibodies, including four mode-1 IGHV3-53 antibodies (blue), two mode-2 IGHV3-53 antibodies (purple), and two IGHV1-2 antibodies (orange). The discrepancy between CV05-163 neutralizing SARS-CoV-2 pseudotyped virus (IC50 = 0.47 μg/ml) and authentic virus (IC50 = 0.02 μg/ml) reported in our previous study (17) is possibly due to different systems (pseudovirus versus authentic virus) and host cells (HeLa cells versus Vero E6 cells) used in these experiments.
” data-icon-position=”” data-hide-link-title=”0″>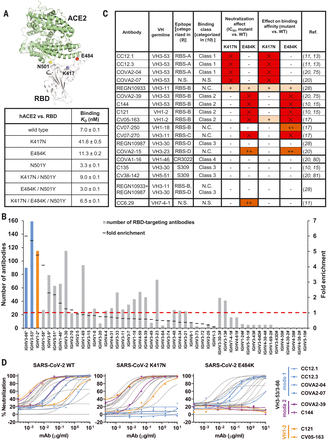

(A) Emergent mutations (spheres) in the RBS of B.1.351 and P.1 lineages are mapped onto a structure of SARS-CoV-2 RBD (white) in complex with ACE2 (green) (PDB 6M0J) (90). Binding affinities of Fc-tagged human ACE2 against SARS-CoV-2 RBD wild type and mutants were assayed by biolayer interferometry (BLI) experiments. Detailed sensorgrams are shown in fig. S1. (B) Distribution of IGHV gene usage. Numbers of RBD-targeting antibodies encoded by each IGHV gene are shown. The frequently used IGHV3-53 and IGHV3-66 genes are highlighted in blue, and IGHV1-2 in orange. The IGHV gene usage in 1593 SARS-CoV-2 RBD-targeting antibodies (11, 14–44) relative to healthy individuals (baseline) (76) (fold enrichment) is shown as black line segments. Hashtags (#) denote IGHV gene frequencies in healthy individuals that were not reported in (76). Asterisks denote IGHV genes that are significantly enriched over the baseline repertoire (76) (P < 0.05, one-sample proportion test with Bonferroni correction). A fold enrichment of 1 (red dashed line) represents no difference over baseline. (C) Effects of single mutations on the neutralization activity and binding affinity of each neutralizing antibody. “–” denotes an increase in half-maximal inhibitory concentration (IC50) or KD by a factor of 100. Results in red with “×” indicate that no neutralization activity or binding was detected at the highest amount of immunoglobulin G used. N.C., not categorized in the original studies; N.S., no structure available. (D) Neutralization of pseudotyped SARS-CoV-2 virus and variants carrying K417N or E484K mutations. We tested a panel of 17 neutralizing antibodies, including four mode-1 IGHV3-53 antibodies (blue), two mode-2 IGHV3-53 antibodies (purple), and two IGHV1-2 antibodies (orange). The discrepancy between CV05-163 neutralizing SARS-CoV-2 pseudotyped virus (IC50 = 0.47 μg/ml) and authentic virus (IC50 = 0.02 μg/ml) reported in our previous study (17) is possibly due to different systems (pseudovirus versus authentic virus) and host cells (HeLa cells versus Vero E6 cells) used in these experiments.
N501Y was previously reported to enhance binding to the human ACE2 receptor (6, 7). Here, we quantified binding of K417N, E484K, N501Y, and double and triple combinations in the RBD to ACE2 by biolayer interferometry (Fig. 1A and fig. S1). N501Y indeed increased RBD binding to ACE2 relative to wild-type RBD (binding affinity KD = 3.3 nM versus 7.0 nM), whereas K417N substantially reduced ACE2 binding (41.6 nM). E484K slightly reduced binding (11.3 nM). N501Y could rescue binding of K417N (9.0 nM), and the triple mutant K417N/E484K/N501Y (as in B.1.351) had similar binding (6.5 nM) to the wild type (Fig. 1A and fig. S1). Consistently, K417N/T mutations are associated with N501Y in naturally circulating SARS-CoV-2. Among 585,054 SARS-CoV-2 genome sequences in the GISAID database (5 March 2021) (8), about 95% of K417N/T mutations occur with N501Y, despite N501Y being present in only 21% of all analyzed sequences. In contrast, only 36% of E484K mutations occur with N501Y.
We and others have shown that most SARS-CoV-2 nAbs that target the RBD and their epitopes can be classified into different sites and subsites (9–12). Certain IGHV genes are highly enriched in the antibody response to SARS-CoV-2 infection, with IGHV3-53 (11, 13–16) and IGHV3-66, which differ by only one conservative substitution (V12I), and IGHV1-2 (11, 17, 18) being the most enriched IGHV genes used among 1593 RBD-targeting antibodies from 32 studies (11, 14–44) (Fig. 1B). We investigated the effects of the prevalent SARS-CoV-2 mutations on neutralization by these major classes of antibodies found in multiple donors, and the consequences for current vaccines and therapeutics.
K417N and E484K in VOCs B.1.351 and P.1 have been reported to decrease the neutralizing activity of sera as well as neutralizing monoclonal antibodies (mAbs) isolated from COVID-19 convalescent plasma and vaccinated individuals (45–62). Relative to wild-type SARS-CoV-2, B.1.351 is more resistant to neutralization by convalescent plasma and mRNA vaccine sera by a factor of ~8 to 14, and P.1 is more resistant by a factor of ~2.6 to 5 (63–68). Some variants are able to escape neutralization by some nAbs (e.g., LY-CoV555, 910-30, COVOX-384, S2H58, C671, etc.), whereas others retain activity (e.g., 1-57, 2-7, mAb-222, S309, S2E12, COV2-2196, C669, etc.) (50, 57, 63, 66, 69, 70). Here, we selected a representative panel of 17 human nAbs isolated from COVID-19 patients or humanized mice to study the escape mechanism. These nAbs cover all known neutralizing sites on the RBD and include those encoded by V genes that are the most frequently used and also significantly enriched (Fig. 1B). We tested the activity of a panel of nAbs against wild-type (Wuhan strain) SARS-CoV-2 pseudovirus and single mutants K417N and E484K (Fig. 1C). Binding and neutralization of four and five antibodies out of the 17 tested were abolished by K417N and E484K, respectively. Strikingly, binding and neutralization by all six highly potent IGHV3-53 antibodies (71) that we tested were abrogated by either K417N (RBS-A/class 1) or E484K (RBS-B/class 2) (Fig. 1, C and D, and fig. S2). In addition, binding and neutralization of IGHV1-2 antibodies was severely reduced for the E484K mutation (Fig. 1, C and D, and fig. S2).
We next examined 54 SARS-CoV-2 RBD-targeting human antibodies with available structures. The antibody epitopes on the RBD can be classified into six sites—four RBS subsites, RBS-A, B, C, and D; CR3022 site; and S309 site (fig. S3) (72)—that are generally related to the four classes assigned in (10) (Fig. 1C). Of 23 IGHV3-53/3-66 antibodies, 21 target RBS-A (Fig. 2A). All IGHV1-2 antibodies with known structures bind to the RBS-B epitope. A large fraction of antibodies in these two main families make contact with Lys417, Glu484, or Asn501 in their epitopes (Fig. 2A) (73). Almost all RBS-A antibodies interact extensively with Lys417 and Asn501, whereas most RBS-B and RBS-C antibodies contact Glu484, and most RBS-C antibodies interact with Leu452. We also examined the buried surface area (BSA) of Lys417, Glu484, and Asn501 upon interaction with these RBD-targeting antibodies (fig. S3C). The extensive BSA confirmed why mutations at positions 417 and 484 affect binding and neutralization. Antibodies targeting RBS-D, or the cross-neutralizing S309 and CR3022 sites, are minimally or not involved in interactions with these four RBD mutations (Fig. 2A and fig. S3C).
(A) Antibodies making contact with RBD residues Lys417, Glu484, and Asn501 are represented by blue, red, and yellow boxes, respectively (cutoff distance = 4 Å). Antibodies encoded by the most frequently elicited IGHV3-53/3-66 and IGHV1-2 in convalescent patients are shown in green and orange boxes, respectively. Antibodies are ordered by epitopes originally classified in (9) with an additional epitope, RBS-D, that maps to a region in the RBS above or slightly overlapping with the S309 site. Details of the epitope classifications are shown in fig. S3A. Structures of RBD-targeting antibodies that were isolated from patients are analyzed (91). (B and C) Residues that are mutated in recently circulating variants are integral to the binding sites of IGHV3-53 antibodies. Representative structures are shown for IGHV3-53 binding mode 1 [CC12.1 (PDB 6XC3), CC12.3 (PDB 6XC4) (13), and COVA2-04 (PDB 7JMO) (75)] (B) and binding mode 2 [COVA2-39 (PDB 7JMP) (75)] (C). The SARS-CoV-2 RBD is in white and Fabs in different colors. Residues Lys417 and Glu484 are represented by blue and red spheres, respectively. Hydrogen bonds and salt bridges are represented by black dashed lines.
” data-icon-position=”” data-hide-link-title=”0″>

(A) Antibodies making contact with RBD residues Lys417, Glu484, and Asn501 are represented by blue, red, and yellow boxes, respectively (cutoff distance = 4 Å). Antibodies encoded by the most frequently elicited IGHV3-53/3-66 and IGHV1-2 in convalescent patients are shown in green and orange boxes, respectively. Antibodies are ordered by epitopes originally classified in (9) with an additional epitope, RBS-D, that maps to a region in the RBS above or slightly overlapping with the S309 site. Details of the epitope classifications are shown in fig. S3A. Structures of RBD-targeting antibodies that were isolated from patients are analyzed (91). (B and C) Residues that are mutated in recently circulating variants are integral to the binding sites of IGHV3-53 antibodies. Representative structures are shown for IGHV3-53 binding mode 1 [CC12.1 (PDB 6XC3), CC12.3 (PDB 6XC4) (13), and COVA2-04 (PDB 7JMO) (75)] (B) and binding mode 2 [COVA2-39 (PDB 7JMP) (75)] (C). The SARS-CoV-2 RBD is in white and Fabs in different colors. Residues Lys417 and Glu484 are represented by blue and red spheres, respectively. Hydrogen bonds and salt bridges are represented by black dashed lines.
IGHV3-53/3-66 RBD antibodies can adopt two different binding modes (9, 10), which we refer here to as binding modes 1 and 2 (74), with distinct epitopes and approach angles (Fig. 2B and fig. S4). All IGHV3-53/3-66 RBD antibodies to date with binding mode 1 have a short, heavy-chain complementarity-determining region 3 (CDR H3) of <15 amino acids (Kabat numbering) and bind RBS-A (13, 16, 32), whereas those with binding mode 2 have a longer CDR H3 (≥15 amino acids) and target RBS-B (9, 10, 75). These dual binding modes enhance recognition of this antibody family for the SARS-CoV-2 RBD, although most IGHV3-53/3-66 RBD antibodies adopt binding mode 1 (Fig. 2 and fig. S4). Lys417 is a key epitope residue for antibodies with IGHV3-53/3-66 binding mode 1 (Fig. 2B and fig. S4). IGHV3-53 germline residues VH Tyr33 and Tyr52 make hydrophobic interactions with the aliphatic moiety of Lys417, and its ε-amino group interacts with CDR H3 through a salt bridge (Asp97 or Glu97), hydrogen bond (H-bond), or cation-π interaction (Phe99) (Fig. 2B). K417N/T would diminish such interactions and therefore affect antibody binding and neutralization, providing a structural explanation for K417N escape in IGHV3-53/3-66 antibodies with binding mode 1 (Fig. 1, C and D, Fig. 2B, and fig. S2). In contrast, IGHV3-53 antibodies with binding mode 2 do not interact with RBD-Lys417 (fig. S4) but with Glu484 through H-bonds with CDR H2 (Fig. 2C). Consistently, binding and neutralization of IGHV3-53 antibodies with binding mode 2 (fig. S4) are abolished by E484K but not K417N (Fig. 1, C and D, and fig. S2). Interestingly, unlike most IGHV3-53 antibodies that are sensitive to K417N/T or E484K, a recently discovered IGHV3-53-encoded mAb-222, which binds RBS, retains activity against P.1 and B.1.351. The mAb-222 light chain could largely restore the neutralization potency of other IGHV3-53 antibodies, which suggests that light-chain interactions can compensate for loss of binding of K417N/T by the heavy chain (63). However, this antibody may represent only a small portion of IGHV3-55/3-66 antibodies that can neutralize VOCs.
Among the IGHV genes used in RBD antibodies, IGHV1-2 is also highly enriched over the baseline frequency in the antibody repertoire of healthy individuals (76) and is second only to IGHV3-53/3-66 (Fig. 1B). We compared three structures of IGHV1-2 antibodies, namely 2-4 (27), S2M11 (30), and C121 (10), that target RBS-B. Despite being encoded by different IGK(L)V genes, 2-4 (IGLV2-8), S2M11 (IGKV3-20), and C121 (IGLV2-23) share a nearly identical binding mode and epitope (Fig. 3A). Structural analysis reveals that the VH Gly26-Tyr-Thr-Phe-Thr-Gly-(Tyr)-Tyr33, Trp50-(Ile)-Asn/Ser-(Pro)-X-Ser-X-Gly-Thr57, and Thr73-Ser-(Ile)-Ser/Thr76 motifs are important for RBD binding (fig. S5, A to D). Although only a small part of the epitope interacts with the light chains of 2-4, S2M11, and C121, VL residues 32 and 91 (and also residue 30 in some antibodies) play an important role in forming a hydrophobic pocket together with VH residues for binding RBD-Phe486, which is another key binding residue in such classes of antibodies (9) (fig. S5, E to I). Three other IGHV1-2 antibodies, 2-43, 2-15, and H4, also bind in a similar mode (77), further highlighting the structural convergence of IGHV1-2 antibodies in targeting the same RBD epitope. All IGHV1-2 antibodies to date form extensive interactions with Glu484 (Fig. 3A and fig. S3C). In particular, germline-encoded VH Tyr33, Asn52 (somatically mutated to Ser52 in C121), and Ser54 are involved in polar interactions with the RBD-Glu484 side chain that would be altered by substitution with Lys (Fig. 3A) and thereby diminish binding and neutralization of IGHV1-2 antibodies against E484K (Fig. 1, C and D, and fig. S2).
(A) Binding mode 1; (B) binding mode 2. Heavy and light chains of antibody 2-4 (PDB 6XEY) (27) are shown in pink and light pink, respectively; S2M11 (PDB 7K43) (30) in orange and yellow; C121 (PDB 7K8X) (10) in dark and light green; and CV05-163 in cyan and light cyan. The RBD is shown in white. Glu484 and Lys417 are highlighted as red and blue spheres, respectively. Hydrogen bonds are represented by dashed lines; hydrogen bonds are not shown for C121 because of limited resolution (3.9 Å).
” data-icon-position=”” data-hide-link-title=”0″>




